PhysioGAN: Training High Fidelity Generative Model for Physiological Sensor Readings
Paper and Code
Apr 25, 2022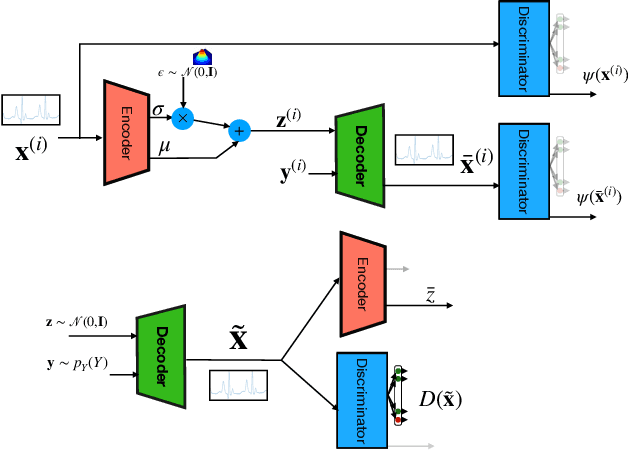
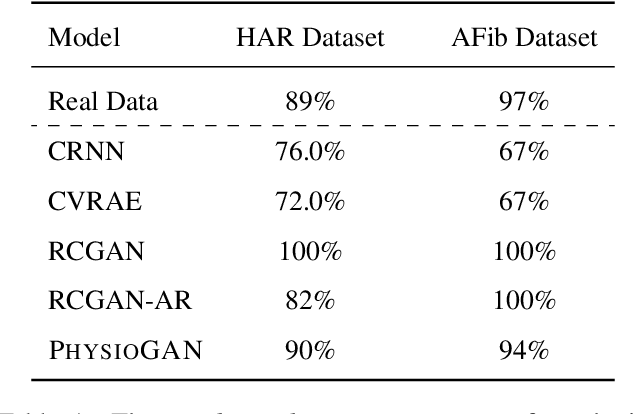
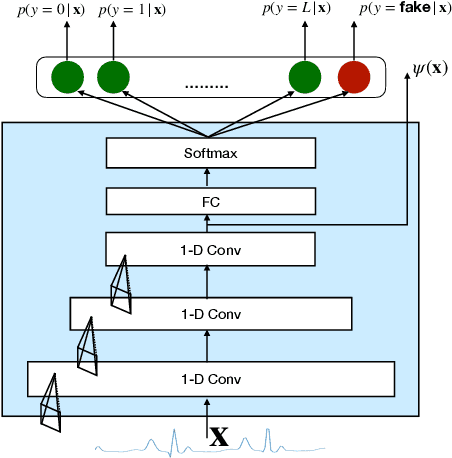
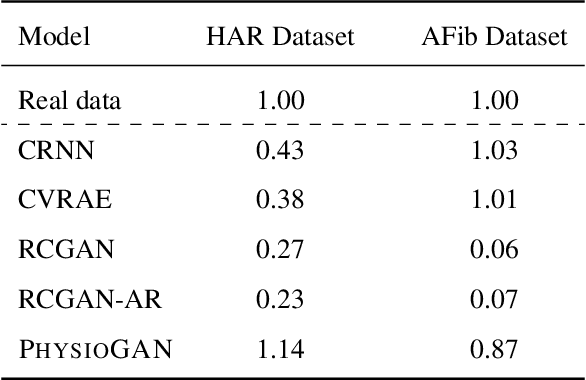
Generative models such as the variational autoencoder (VAE) and the generative adversarial networks (GAN) have proven to be incredibly powerful for the generation of synthetic data that preserves statistical properties and utility of real-world datasets, especially in the context of image and natural language text. Nevertheless, until now, there has no successful demonstration of how to apply either method for generating useful physiological sensory data. The state-of-the-art techniques in this context have achieved only limited success. We present PHYSIOGAN, a generative model to produce high fidelity synthetic physiological sensor data readings. PHYSIOGAN consists of an encoder, decoder, and a discriminator. We evaluate PHYSIOGAN against the state-of-the-art techniques using two different real-world datasets: ECG classification and activity recognition from motion sensors datasets. We compare PHYSIOGAN to the baseline models not only the accuracy of class conditional generation but also the sample diversity and sample novelty of the synthetic datasets. We prove that PHYSIOGAN generates samples with higher utility than other generative models by showing that classification models trained on only synthetic data generated by PHYSIOGAN have only 10% and 20% decrease in their classification accuracy relative to classification models trained on the real data. Furthermore, we demonstrate the use of PHYSIOGAN for sensor data imputation in creating plausible results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge