Phase retrieval from 4-dimensional electron diffraction datasets
Paper and Code
Jun 15, 2021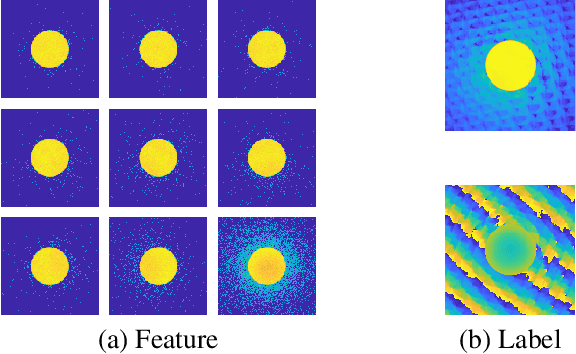
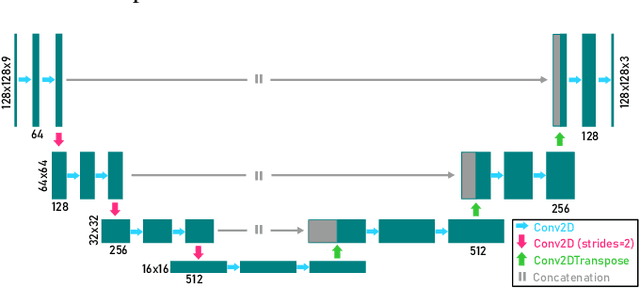

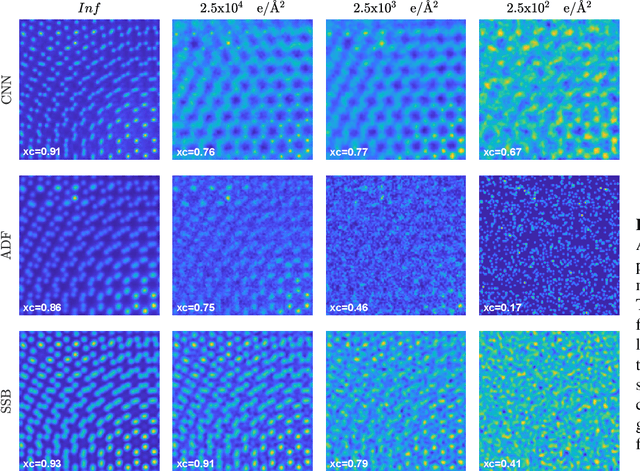
We present a computational imaging mode for large scale electron microscopy data, which retrieves a complex wave from noisy/sparse intensity recordings using a deep learning approach and subsequently reconstructs an image of the specimen from the Convolutional Neural Network (CNN) predicted exit waves. We demonstrate that an appropriate forward model in combination with open data frameworks can be used to generate large synthetic datasets for training. In combination with augmenting the data with Poisson noise corresponding to varying dose-values, we effectively eliminate overfitting issues. The U-NET based architecture of the CNN is adapted to the task at hand and performs well while maintaining a relatively small size and fast performance. The validity of the approach is confirmed by comparing the reconstruction to well-established methods using simulated, as well as real electron microscopy data. The proposed method is shown to be effective particularly in the low dose range, evident by strong suppression of noise, good spatial resolution, and sensitivity to different atom types, enabling the simultaneous visualisation of light and heavy elements and making different atomic species distinguishable. Since the method acts on a very local scale and is comparatively fast it bears the potential to be used for near-real-time reconstruction during data acquisition.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge