Multi-Modal and Multi-Factor Branching Time Active Inference
Paper and Code
Jun 24, 2022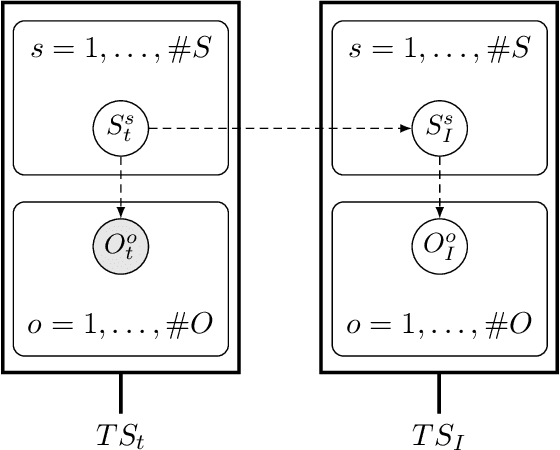
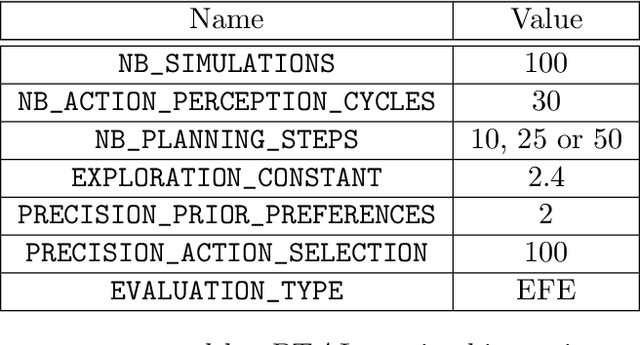

Active inference is a state-of-the-art framework for modelling the brain that explains a wide range of mechanisms such as habit formation, dopaminergic discharge and curiosity. Recently, two versions of branching time active inference (BTAI) based on Monte-Carlo tree search have been developed to handle the exponential (space and time) complexity class that occurs when computing the prior over all possible policies up to the time horizon. However, those two versions of BTAI still suffer from an exponential complexity class w.r.t the number of observed and latent variables being modelled. In the present paper, we resolve this limitation by first allowing the modelling of several observations, each of them having its own likelihood mapping. Similarly, we allow each latent state to have its own transition mapping. The inference algorithm then exploits the factorisation of the likelihood and transition mappings to accelerate the computation of the posterior. Those two optimisations were tested on the dSprites environment in which the metadata of the dSprites dataset was used as input to the model instead of the dSprites images. On this task, $BTAI_{VMP}$ (Champion et al., 2022b,a) was able to solve 96.9\% of the task in 5.1 seconds, and $BTAI_{BF}$ (Champion et al., 2021a) was able to solve 98.6\% of the task in 17.5 seconds. Our new approach ($BTAI_{3MF}$) outperformed both of its predecessors by solving the task completly (100\%) in only 2.559 seconds. Finally, $BTAI_{3MF}$ has been implemented in a flexible and easy to use (python) package, and we developed a graphical user interface to enable the inspection of the model's beliefs, planning process and behaviour.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge