Molecular Classification Using Hyperdimensional Graph Classification
Paper and Code
Mar 18, 2024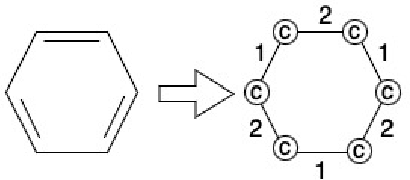
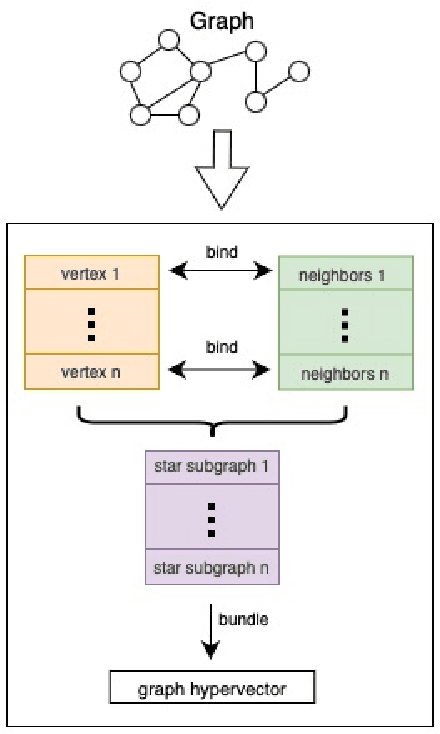
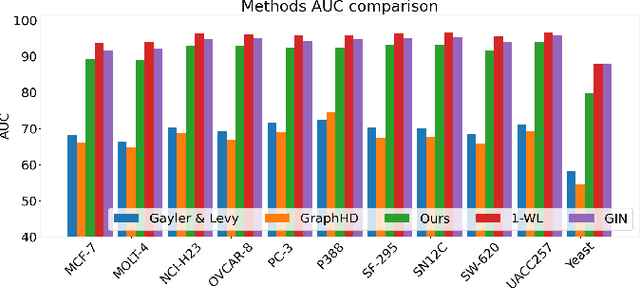
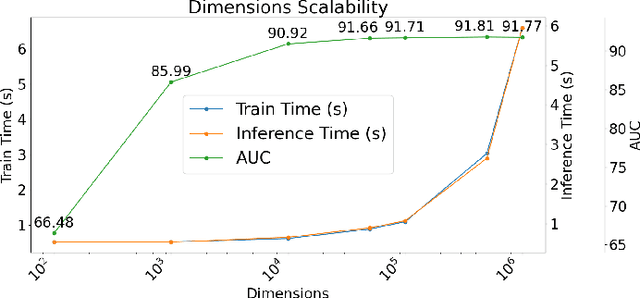
Our work introduces an innovative approach to graph learning by leveraging Hyperdimensional Computing. Graphs serve as a widely embraced method for conveying information, and their utilization in learning has gained significant attention. This is notable in the field of chemoinformatics, where learning from graph representations plays a pivotal role. An important application within this domain involves the identification of cancerous cells across diverse molecular structures. We propose an HDC-based model that demonstrates comparable Area Under the Curve results when compared to state-of-the-art models like Graph Neural Networks (GNNs) or the Weisfieler-Lehman graph kernel (WL). Moreover, it outperforms previously proposed hyperdimensional computing graph learning methods. Furthermore, it achieves noteworthy speed enhancements, boasting a 40x acceleration in the training phase and a 15x improvement in inference time compared to GNN and WL models. This not only underscores the efficacy of the HDC-based method, but also highlights its potential for expedited and resource-efficient graph learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge