MoFlow: An Invertible Flow Model for Generating Molecular Graphs
Paper and Code
Jun 17, 2020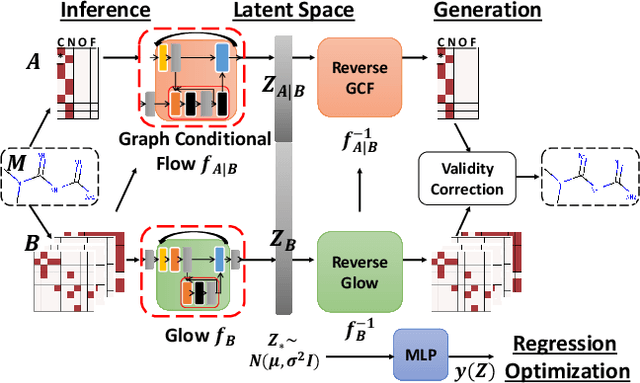
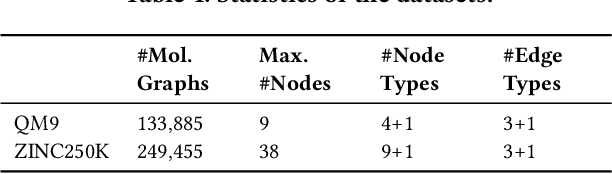
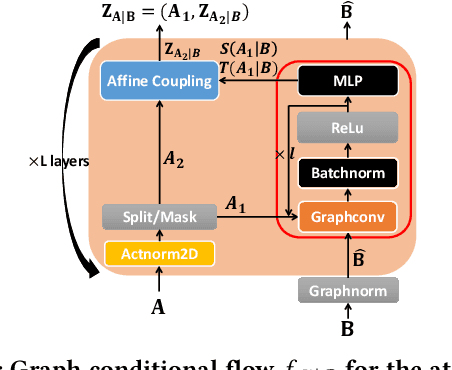
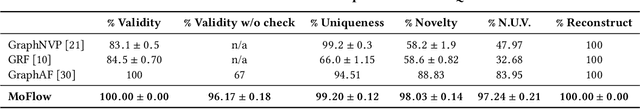
Generating molecular graphs with desired chemical properties driven by deep graph generative models provides a very promising way to accelerate drug discovery process. Such graph generative models usually consist of two steps: learning latent representations and generation of molecular graphs. However, to generate novel and chemically-valid molecular graphs from latent representations is very challenging because of the chemical constraints and combinatorial complexity of molecular graphs. In this paper, we propose MoFlow, a flow-based graph generative model to learn invertible mappings between molecular graphs and their latent representations. To generate molecular graphs, our MoFlow first generates bonds (edges) through a Glow based model, then generates atoms (nodes) given bonds by a novel graph conditional flow, and finally assembles them into a chemically valid molecular graph with a posthoc validity correction. Our MoFlow has merits including exact and tractable likelihood training, efficient one-pass embedding and generation, chemical validity guarantees, 100\% reconstruction of training data, and good generalization ability. We validate our model by four tasks: molecular graph generation and reconstruction, visualization of the continuous latent space, property optimization, and constrained property optimization. Our MoFlow achieves state-of-the-art performance, which implies its potential efficiency and effectiveness to explore large chemical space for drug discovery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge