Learning a Generative Model of Cancer Metastasis
Paper and Code
Jan 17, 2019
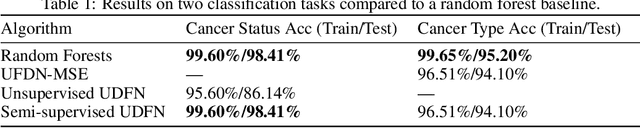
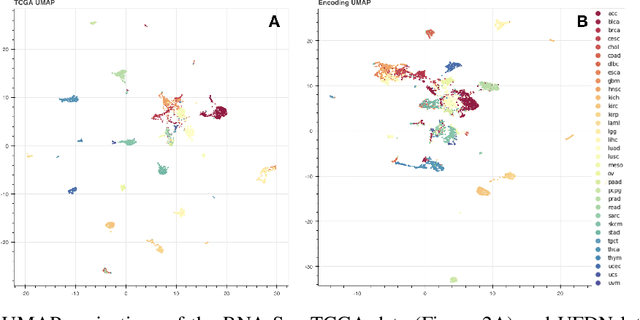
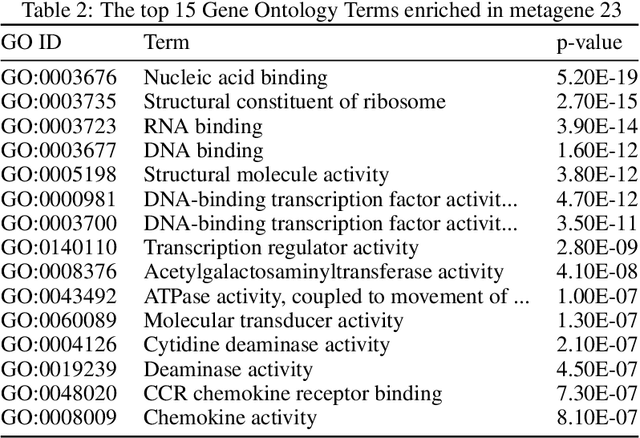
We introduce a Unified Disentanglement Network (UFDN) trained on The Cancer Genome Atlas (TCGA). We demonstrate that the UFDN learns a biologically relevant latent space of gene expression data by applying our network to two classification tasks of cancer status and cancer type. Our UFDN specific algorithms perform comparably to random forest methods. The UFDN allows for continuous, partial interpolation between distinct cancer types. Furthermore, we perform an analysis of differentially expressed genes between skin cutaneous melanoma(SKCM) samples and the same samples interpolated into glioblastoma (GBM). We demonstrate that our interpolations learn relevant metagenes that recapitulate known glioblastoma mechanisms and suggest possible starting points for investigations into the metastasis of SKCM into GBM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge