Learning a Continuous Representation of 3D Molecular Structures with Deep Generative Models
Paper and Code
Oct 20, 2020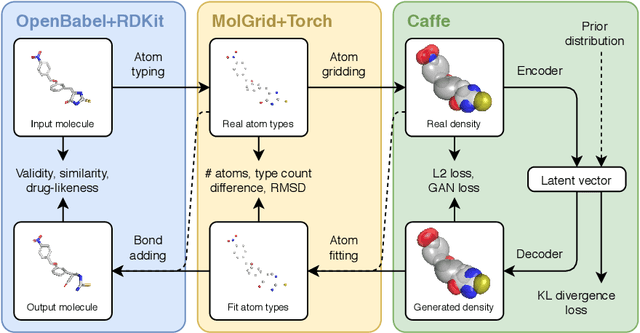
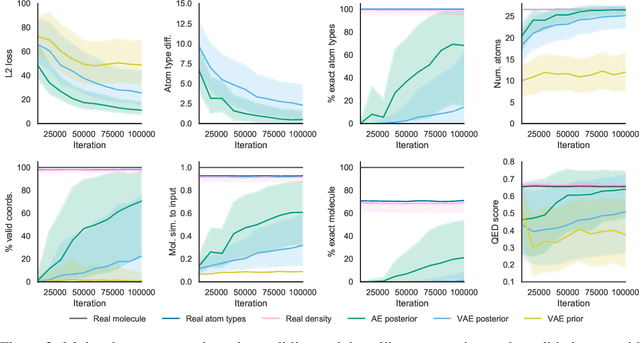
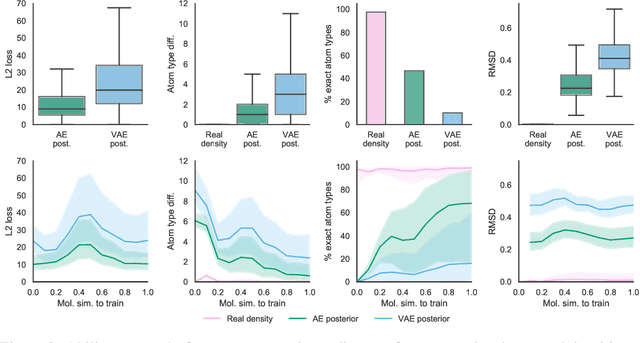
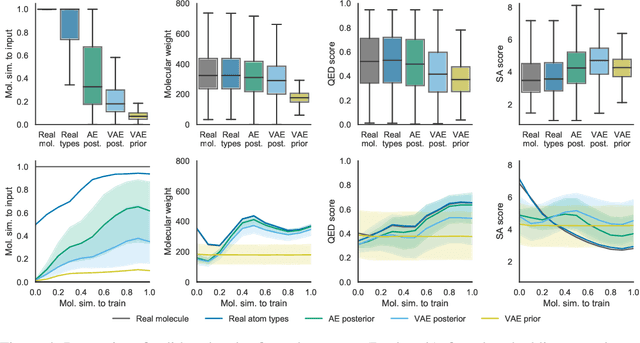
Machine learning methods in drug discovery have primarily focused on virtual screening of molecular libraries using discriminative models. Generative models are an entirely different approach to drug discovery that learn to represent and optimize molecules in a continuous latent space. These methods have already been applied with increasing success to the generation of two dimensional molecules as SMILES strings and molecular graphs. In this work, we describe deep generative models for three dimensional molecular structures using atomic density grids and a novel fitting algorithm that converts continuous grids to discrete molecular structures. Our models jointly represent drug-like molecules and their conformations in a latent space that can be explored through interpolation. We are able to sample diverse sets of molecules based on a given input compound and increase the probability of creating a valid, drug-like molecule.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge