Large-scale biometry with interpretable neural network regression on UK Biobank body MRI
Paper and Code
Mar 10, 2020
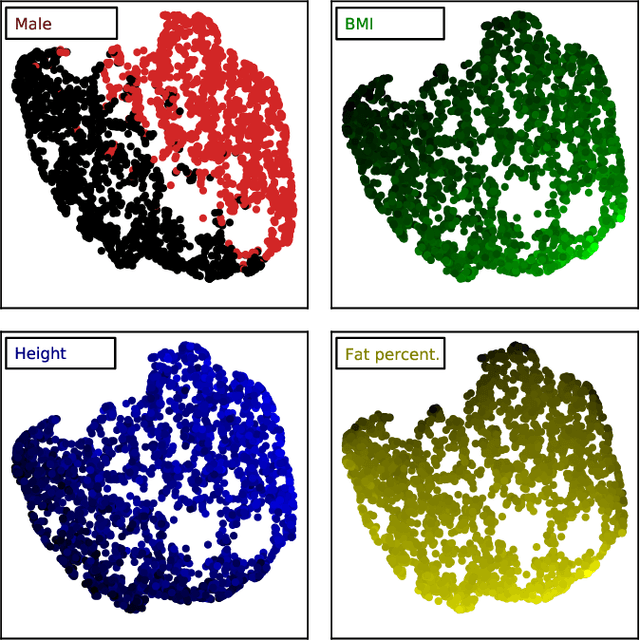
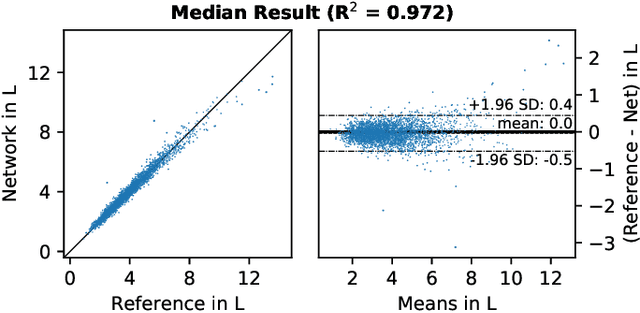
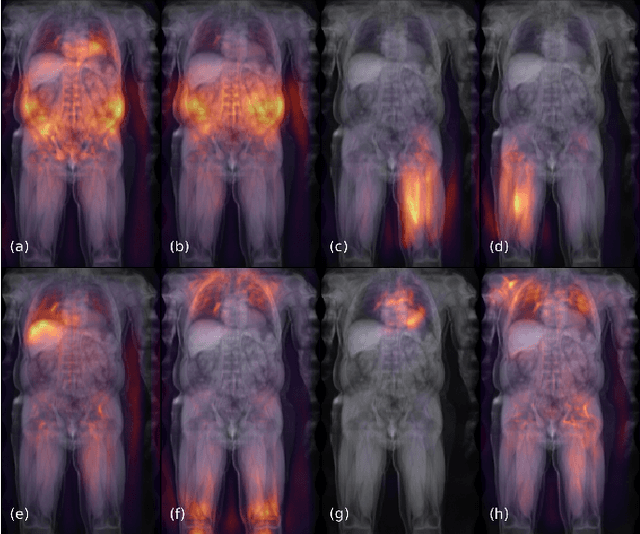
Objective: Automated analysis of MRI with deep regression has the potential to provide medical research with a wide range of biological metrics, inferred at high speed and accuracy. Methods: The UK Biobank study has successfully imaged more than 32,000 volunteer participants with neck-to-knee body MRI. Each scan is linked to extensive metadata, providing a comprehensive survey of imaged anatomy and related health states. Despite its potential for research, this vast amount of data presents a challenge to established methods of evaluation, which often rely on manual input. In this work, neural networks were trained for regression to infer various biological metrics from the neck-to-knee body MRI automatically, with a ResNet50 in 7-fold cross-validation. No manual intervention or ground truth segmentations are required for training. The examined fields span 64 variables derived from anthropometric measurements, dual-energy X-ray absorptiometry (DXA), atlas-based segmentations, and dedicated liver scans. Results: The standardized framework achieved a close fit to the target values (median R^2 > 0.97). Interpretation of aggregated saliency maps indicates that the network correctly targets specific body regions and limbs, and learned to emulate different modalities. On several body composition metrics, the quality of the predictions is within the range of variability observed between established gold standard techniques. Conclusion and Significance: The deep regression framework robustly inferred a wide range of medically relevant metrics from the image data. In practice, this technique could provide accurate, image-based measurements for medical research months or years before the more established reference methods have been fully applied.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge