$l_{1-2}$ GLasso: $L_{1-2}$ Regularized Multi-task Graphical Lasso for Joint Estimation of eQTL Mapping and Gene Network
Paper and Code
Jan 04, 2023

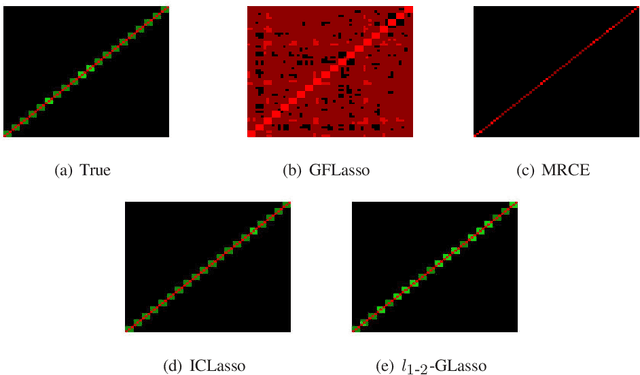

A critical problem in genetics is to discover how gene expression is regulated within cells. Two major tasks of regulatory association learning are : (i) identifying SNP-gene relationships, known as eQTL mapping, and (ii) determining gene-gene relationships, known as gene network estimation. To share information between these two tasks, we focus on the unified model for joint estimation of eQTL mapping and gene network, and propose a $L_{1-2}$ regularized multi-task graphical lasso, named $L_{1-2}$ GLasso. Numerical experiments on artificial datasets demonstrate the competitive performance of $L_{1-2}$ GLasso on capturing the true sparse structure of eQTL mapping and gene network. $L_{1-2}$ GLasso is further applied to real dataset of ADNI-1 and experimental results show that $L_{1 -2}$ GLasso can obtain sparser and more accurate solutions than other commonly-used methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge