Kernel-based retrieval models for hyperspectral image data optimized with Kernel Flows
Paper and Code
Nov 12, 2024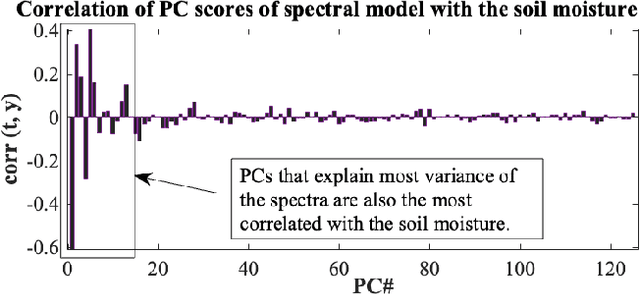
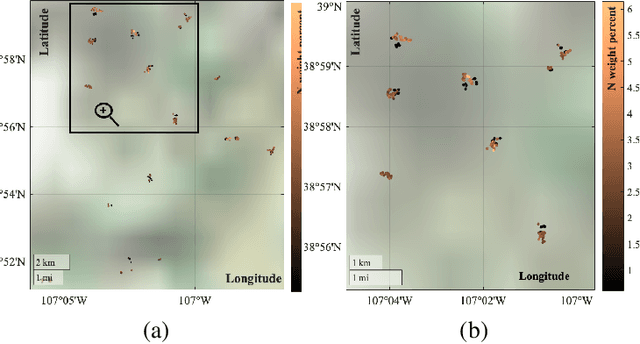
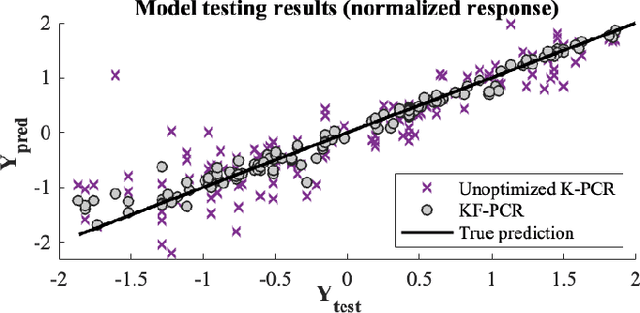
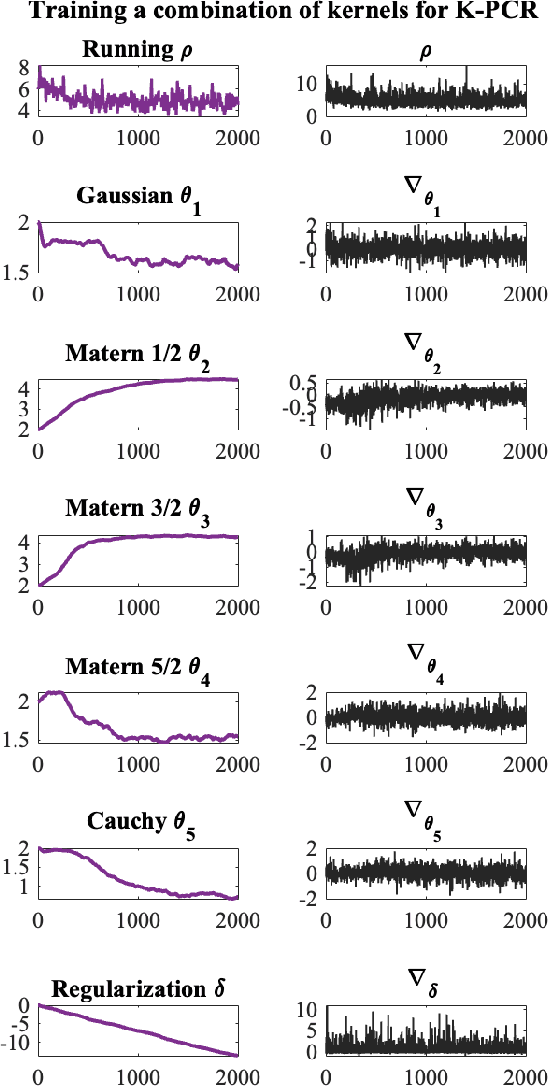
Kernel-based statistical methods are efficient, but their performance depends heavily on the selection of kernel parameters. In literature, the optimization studies on kernel-based chemometric methods is limited and often reduced to grid searching. Previously, the authors introduced Kernel Flows (KF) to learn kernel parameters for Kernel Partial Least-Squares (K-PLS) regression. KF is easy to implement and helps minimize overfitting. In cases of high collinearity between spectra and biogeophysical quantities in spectroscopy, simpler methods like Principal Component Regression (PCR) may be more suitable. In this study, we propose a new KF-type approach to optimize Kernel Principal Component Regression (K-PCR) and test it alongside KF-PLS. Both methods are benchmarked against non-linear regression techniques using two hyperspectral remote sensing datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge