Jansen-MIDAS: a multi-level photomicrograph segmentation software based on isotropic undecimated wavelets
Paper and Code
May 25, 2016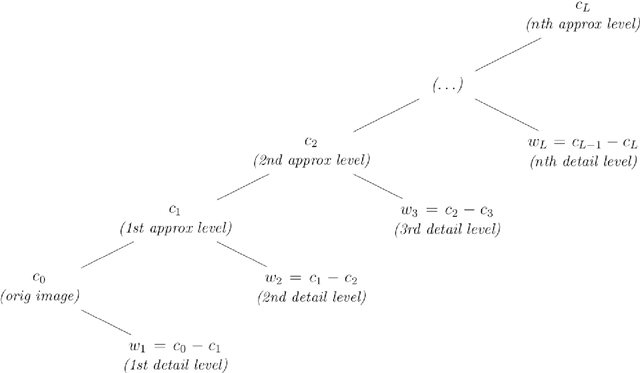
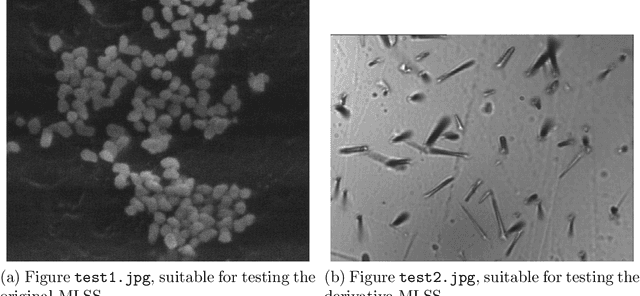
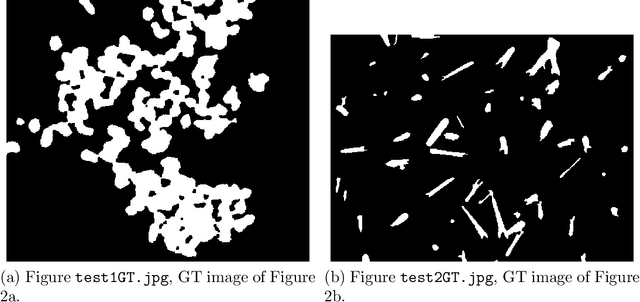
Image segmentation, the process of separating the elements within an image, is frequently used for obtaining information from photomicrographs. However, segmentation methods should be used with reservations: incorrect segmentation can mislead when interpreting regions of interest (ROI), thus decreasing the success rate of additional procedures. Multi-Level Starlet Segmentation (MLSS) and Multi-Level Starlet Optimal Segmentation (MLSOS) were developed to address the photomicrograph segmentation deficiency on general tools. These methods gave rise to Jansen-MIDAS, an open-source software which a scientist can use to obtain a multi-level threshold segmentation of his/hers photomicrographs. This software is presented in two versions: a text-based version, for GNU Octave, and a graphical user interface (GUI) version, for MathWorks MATLAB. It can be used to process several types of images, becoming a reliable alternative to the scientist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge