Investigating Manifold Neighborhood size for Nonlinear Analysis of LIBS Amino Acid Spectra
Paper and Code
May 25, 2021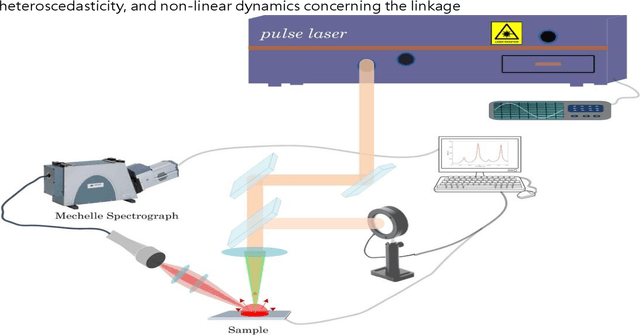
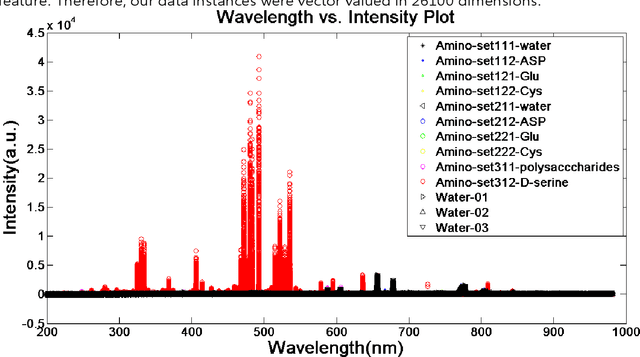
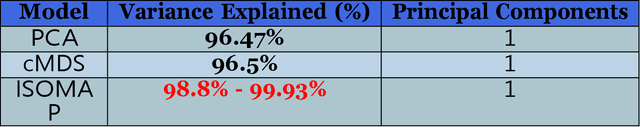
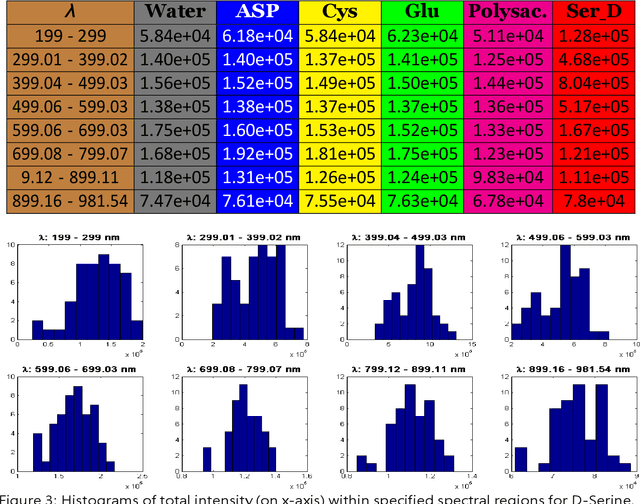
Classification and identification of amino acids in aqueous solutions is important in the study of biomacromolecules. Laser Induced Breakdown Spectroscopy (LIBS) uses high energy laser-pulses for ablation of chemical compounds whose radiated spectra are captured and recorded to reveal molecular structure. Spectral peaks and noise from LIBS are impacted by experimental protocols. Current methods for LIBS spectral analysis achieves promising results using PCA, a linear method. It is well-known that the underlying physical processes behind LIBS are highly nonlinear. Our work set out to understand the impact of LIBS spectra on suitable neighborhood size over which to consider pattern phenomena, if nonlinear methods capture pattern phenomena with increased efficacy, and how they improve classification and identification of compounds. We analyzed four amino acids, polysaccharide, and a control group, water. We developed an information theoretic method for measurement of LIBS energy spectra, implemented manifold methods for nonlinear dimensionality reduction, and found while clustering results were not statistically significantly different, nonlinear methods lead to increased classification accuracy. Moreover, our approach uncovered the contribution of micro-wells (experimental protocol) in LIBS spectra. To the best of our knowledge, ours is the first application of Manifold methods to LIBS amino-acid analysis in the research literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge