Improving ICD-based semantic similarity by accounting for varying degrees of comorbidity
Paper and Code
Aug 14, 2023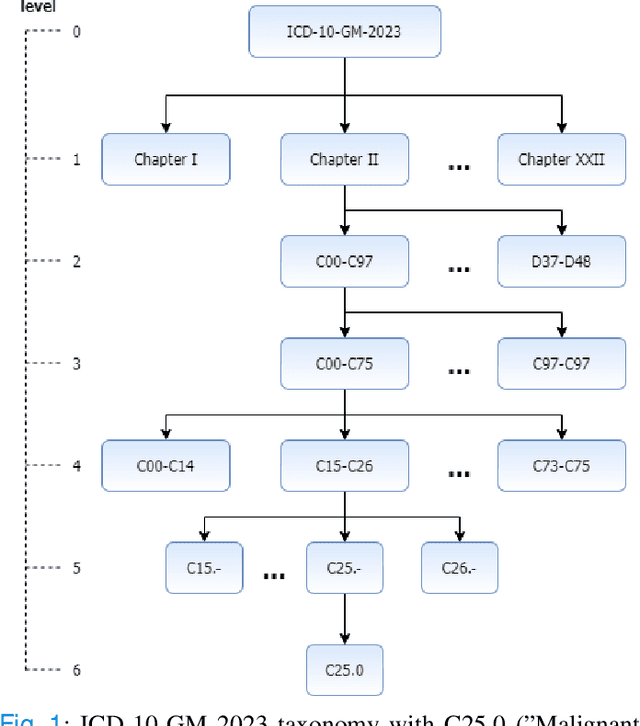
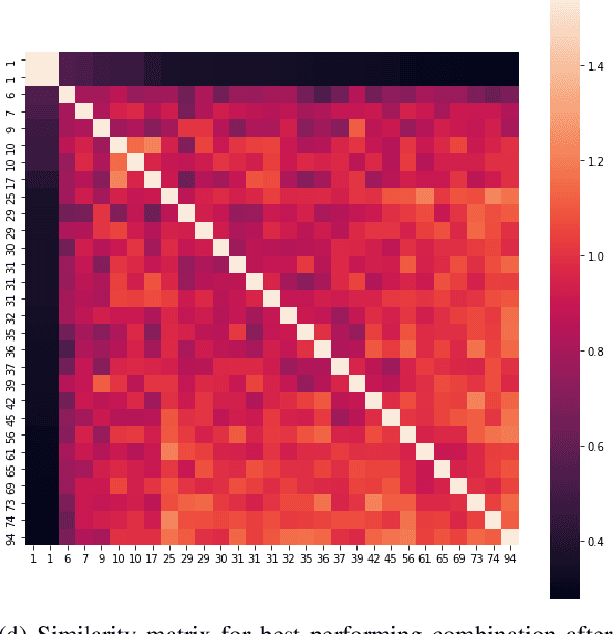
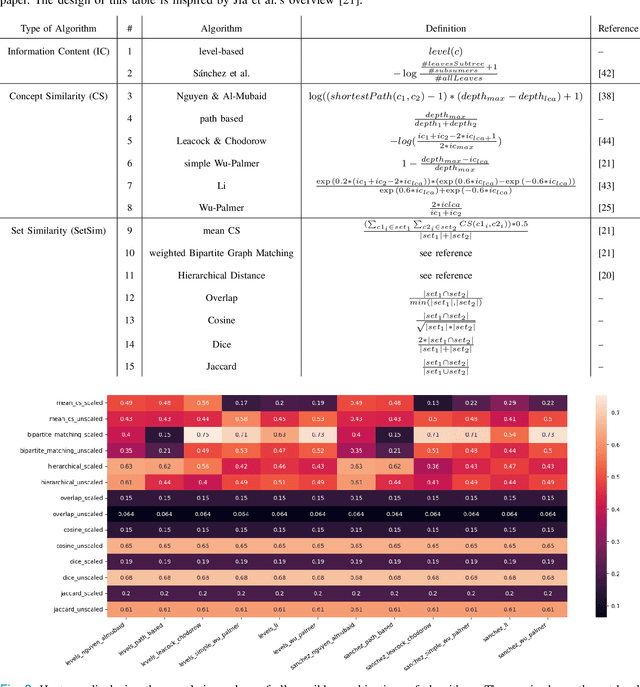
Finding similar patients is a common objective in precision medicine, facilitating treatment outcome assessment and clinical decision support. Choosing widely-available patient features and appropriate mathematical methods for similarity calculations is crucial. International Statistical Classification of Diseases and Related Health Problems (ICD) codes are used worldwide to encode diseases and are available for nearly all patients. Aggregated as sets consisting of primary and secondary diagnoses they can display a degree of comorbidity and reveal comorbidity patterns. It is possible to compute the similarity of patients based on their ICD codes by using semantic similarity algorithms. These algorithms have been traditionally evaluated using a single-term expert rated data set. However, real-word patient data often display varying degrees of documented comorbidities that might impair algorithm performance. To account for this, we present a scale term that considers documented comorbidity-variance. In this work, we compared the performance of 80 combinations of established algorithms in terms of semantic similarity based on ICD-code sets. The sets have been extracted from patients with a C25.X (pancreatic cancer) primary diagnosis and provide a variety of different combinations of ICD-codes. Using our scale term we yielded the best results with a combination of level-based information content, Leacock & Chodorow concept similarity and bipartite graph matching for the set similarities reaching a correlation of 0.75 with our expert's ground truth. Our results highlight the importance of accounting for comorbidity variance while demonstrating how well current semantic similarity algorithms perform.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge