GOLLuM: Gaussian Process Optimized LLMs -- Reframing LLM Finetuning through Bayesian Optimization
Paper and Code
Apr 09, 2025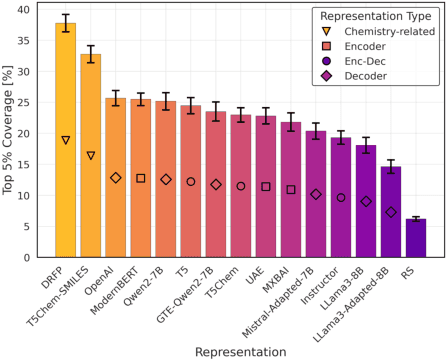
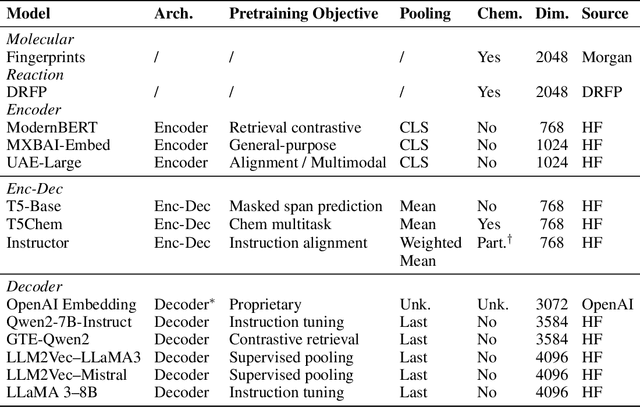
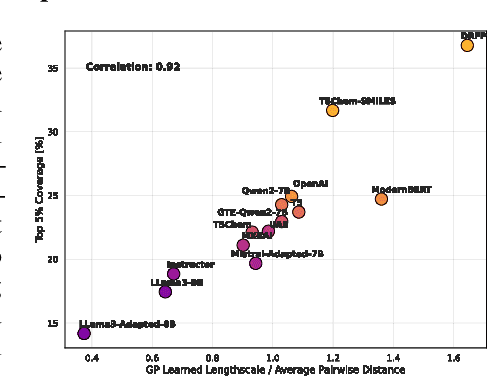
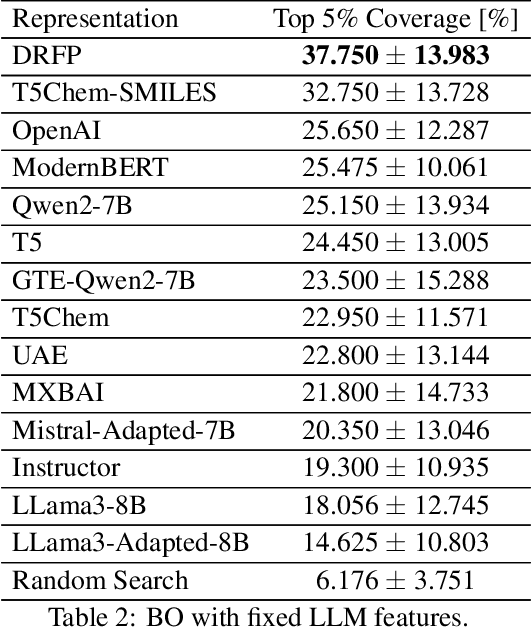
Large Language Models (LLMs) can encode complex relationships in their latent spaces, yet harnessing them for optimization under uncertainty remains challenging. We address this gap with a novel architecture that reframes LLM finetuning as Gaussian process (GP) marginal likelihood optimization via deep kernel methods. We introduce LLM-based deep kernels, jointly optimized with GPs to preserve the benefits of both - LLMs to provide a rich and flexible input space for Bayesian optimization and - GPs to model this space with predictive uncertainty for more efficient sampling. Applied to Buchwald-Hartwig reaction optimization, our method nearly doubles the discovery rate of high-performing reactions compared to static LLM embeddings (from 24% to 43% coverage of the top 5% reactions in just 50 optimization iterations). We also observe a 14% improvement over domain-specific representations without requiring specialized features. Extensive empirical evaluation across 19 benchmarks - ranging from general chemistry to reaction and molecular property optimization - demonstrates our method's robustness, generality, and consistent improvements across: (1) tasks, (2) LLM architectures (encoder, decoder, encoder-decoder), (3) pretraining domains (chemistry-related or general-purpose) and (4) hyperparameter settings (tuned once on a single dataset). Finally, we explain these improvements: joint LLM-GP optimization through marginal likelihood implicitly performs contrastive learning, aligning representations to produce (1) better-structured embedding spaces, (2) improved uncertainty calibration, and (3) more efficient sampling - without requiring any external loss. This work provides both practical advances in sample-efficient optimization and insights into what makes effective Bayesian optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge