Generalized Similarity U: A Non-parametric Test of Association Based on Similarity
Paper and Code
Jan 04, 2018
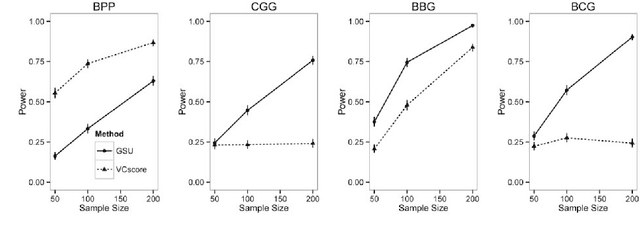
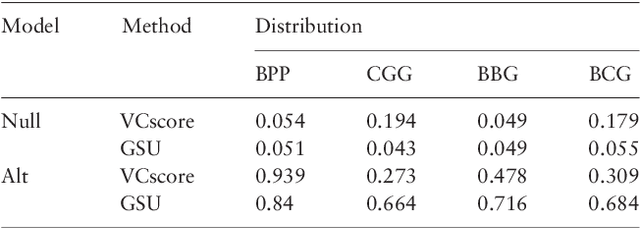

Second generation sequencing technologies are being increasingly used for genetic association studies, where the main research interest is to identify sets of genetic variants that contribute to various phenotype. The phenotype can be univariate disease status, multivariate responses and even high-dimensional outcomes. Considering the genotype and phenotype as two complex objects, this also poses a general statistical problem of testing association between complex objects. We here proposed a similarity-based test, generalized similarity U (GSU), that can test the association between complex objects. We first studied the theoretical properties of the test in a general setting and then focused on the application of the test to sequencing association studies. Based on theoretical analysis, we proposed to use Laplacian kernel based similarity for GSU to boost power and enhance robustness. Through simulation, we found that GSU did have advantages over existing methods in terms of power and robustness. We further performed a whole genome sequencing (WGS) scan for Alzherimer Disease Neuroimaging Initiative (ADNI) data, identifying three genes, APOE, APOC1 and TOMM40, associated with imaging phenotype. We developed a C++ package for analysis of whole genome sequencing data using GSU. The source codes can be downloaded at https://github.com/changshuaiwei/gsu.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge