Generalized knowledge-enhanced framework for biomedical entity and relation extraction
Paper and Code
Aug 13, 2024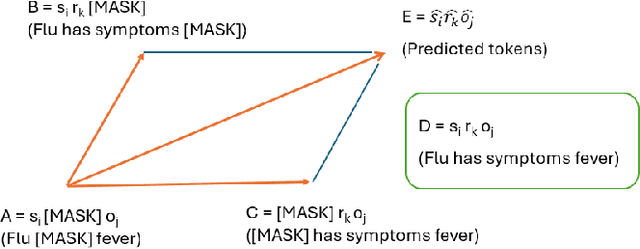
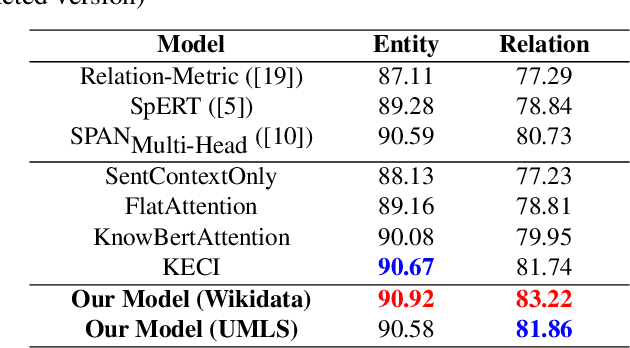
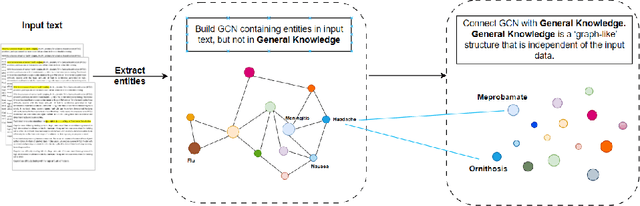
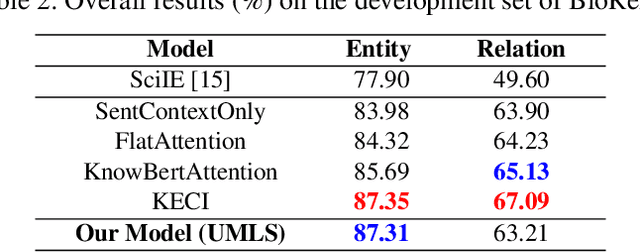
In recent years, there has been an increasing number of frameworks developed for biomedical entity and relation extraction. This research effort aims to address the accelerating growth in biomedical publications and the intricate nature of biomedical texts, which are written for mainly domain experts. To handle these challenges, we develop a novel framework that utilizes external knowledge to construct a task-independent and reusable background knowledge graph for biomedical entity and relation extraction. The design of our model is inspired by how humans learn domain-specific topics. In particular, humans often first acquire the most basic and common knowledge regarding a field to build the foundational knowledge and then use that as a basis for extending to various specialized topics. Our framework employs such common-knowledge-sharing mechanism to build a general neural-network knowledge graph that is learning transferable to different domain-specific biomedical texts effectively. Experimental evaluations demonstrate that our model, equipped with this generalized and cross-transferable knowledge base, achieves competitive performance benchmarks, including BioRelEx for binding interaction detection and ADE for Adverse Drug Effect identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge