Generalizable Protein Interface Prediction with End-to-End Learning
Paper and Code
Sep 21, 2018

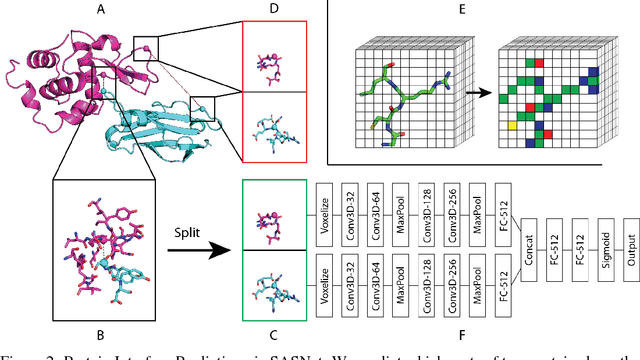
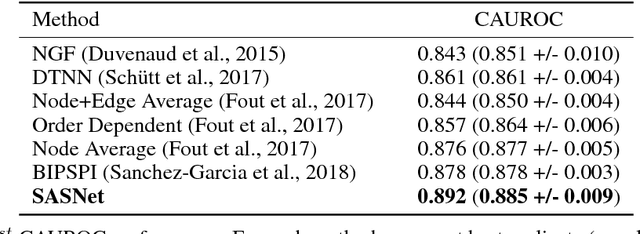
Predicting how proteins interact with one another - that is, which surfaces of one protein bind to which surfaces of another protein - is a central problem in biology. Here we present Siamese Atomic Surfacelet Network (SASNet), the first end-to-end learning method for protein interface prediction. Despite using only spatial coordinates and identities of atoms as inputs, SASNet outperforms state-of-the-art methods that rely on complex, hand-selected features. These results are particularly striking because we train the method entirely on a significantly biased data set that does not account for the fact that proteins deform when binding to one another. Nonetheless, our network maintains high performance, without retraining, when tested on real cases in which proteins do deform. This suggests that it has learned fundamental properties of protein structure and dynamics, which has important implications for a variety of key problems related to biomolecular structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge