Feature versus Raw Sequence: Deep Learning Comparative Study on Predicting Pre-miRNA
Paper and Code
Oct 17, 2017

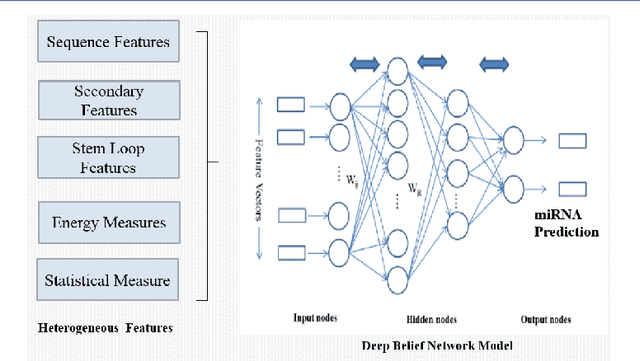

Should we input known genome sequence features or input sequence itself in deep learning framework? As deep learning more popular in various applications, researchers often come to question whether to generate features or use raw sequences for deep learning. To answer this question, we study the prediction accuracy of precursor miRNA prediction of feature-based deep belief network and sequence-based convolution neural network. Tested on a variant of six-layer convolution neural net and three-layer deep belief network, we find the raw sequence input based convolution neural network model performs similar or slightly better than feature based deep belief networks with best accuracy values of 0.995 and 0.990, respectively. Both the models outperform existing benchmarks models. The results shows us that if provided large enough data, well devised raw sequence based deep learning models can replace feature based deep learning models. However, construction of well behaved deep learning model can be very challenging. In cased features can be easily extracted, feature-based deep learning models may be a better alternative.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge