Feature vector regularization in machine learning
Paper and Code
Dec 30, 2013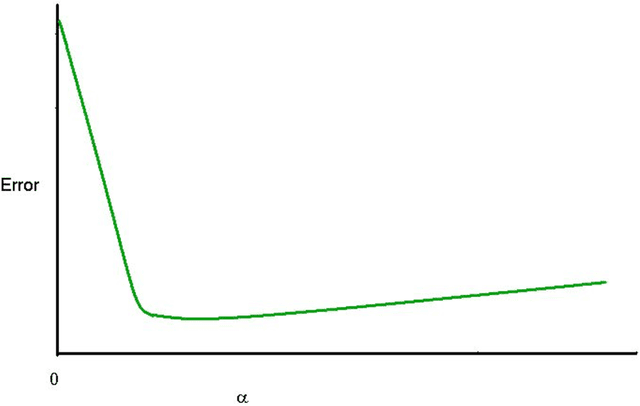
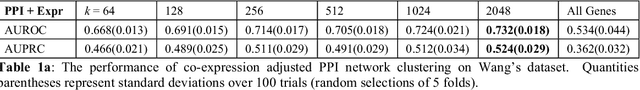
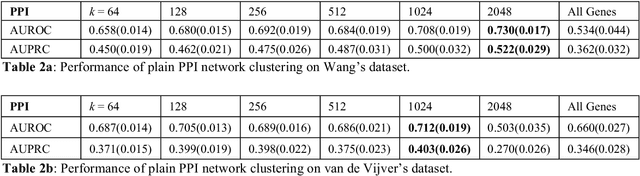
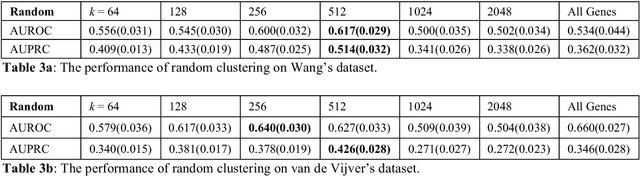
Problems in machine learning (ML) can involve noisy input data, and ML classification methods have reached limiting accuracies when based on standard ML data sets consisting of feature vectors and their classes. Greater accuracy will require incorporation of prior structural information on data into learning. We study methods to regularize feature vectors (unsupervised regularization methods), analogous to supervised regularization for estimating functions in ML. We study regularization (denoising) of ML feature vectors using Tikhonov and other regularization methods for functions on ${\bf R}^n$. A feature vector ${\bf x}=(x_1,\ldots,x_n)=\{x_q\}_{q=1}^n$ is viewed as a function of its index $q$, and smoothed using prior information on its structure. This can involve a penalty functional on feature vectors analogous to those in statistical learning, or use of proximity (e.g. graph) structure on the set of indices. Such feature vector regularization inherits a property from function denoising on ${\bf R}^n$, in that accuracy is non-monotonic in the denoising (regularization) parameter $\alpha$. Under some assumptions about the noise level and the data structure, we show that the best reconstruction accuracy also occurs at a finite positive $\alpha$ in index spaces with graph structures. We adapt two standard function denoising methods used on ${\bf R}^n$, local averaging and kernel regression. In general the index space can be any discrete set with a notion of proximity, e.g. a metric space, a subset of ${\bf R}^n$, or a graph/network, with feature vectors as functions with some notion of continuity. We show this improves feature vector recovery, and thus the subsequent classification or regression done on them. We give an example in gene expression analysis for cancer classification with the genome as an index space and network structure based protein-protein interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge