Exploration of Rashomon Set Assists Explanations for Medical Data
Paper and Code
Aug 22, 2023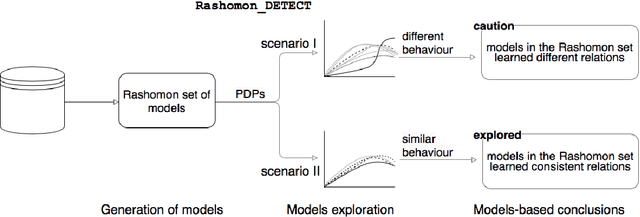
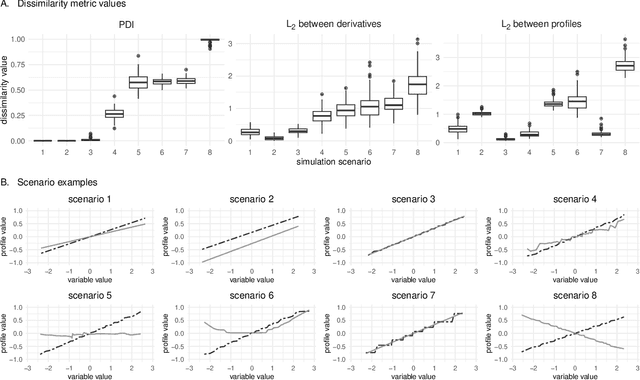
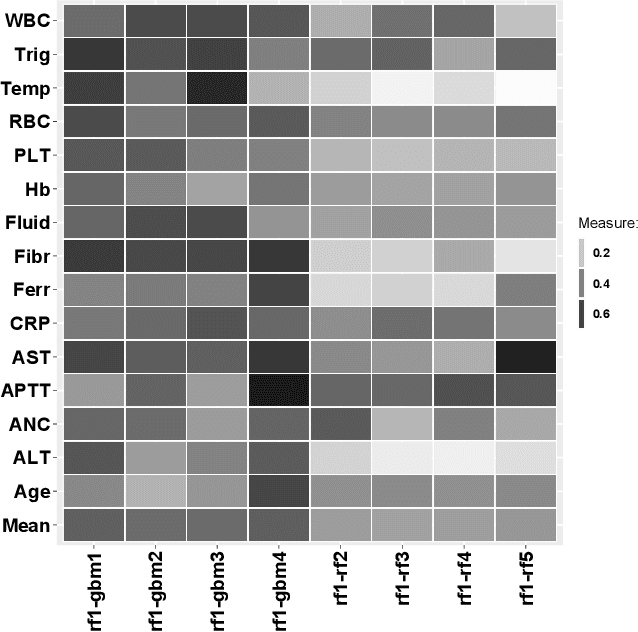
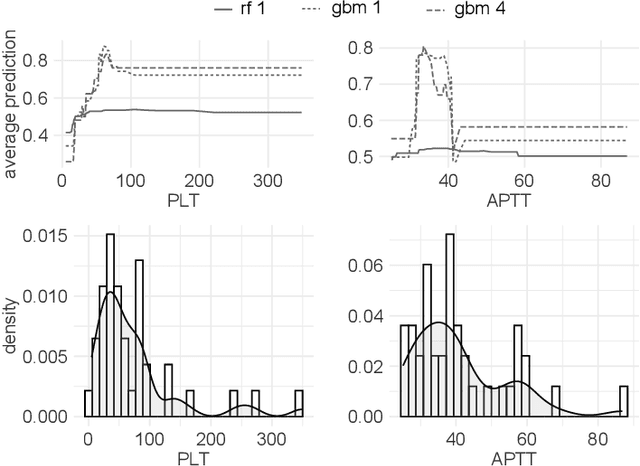
The machine learning modeling process conventionally culminates in selecting a single model that maximizes a selected performance metric. However, this approach leads to abandoning a more profound analysis of slightly inferior models. Particularly in medical and healthcare studies, where the objective extends beyond predictions to valuable insight generation, relying solely on performance metrics can result in misleading or incomplete conclusions. This problem is particularly pertinent when dealing with a set of models with performance close to maximum one, known as $\textit{Rashomon set}$. Such a set can be numerous and may contain models describing the data in a different way, which calls for comprehensive analysis. This paper introduces a novel process to explore Rashomon set models, extending the conventional modeling approach. The cornerstone is the identification of the most different models within the Rashomon set, facilitated by the introduced $\texttt{Rashomon_DETECT}$ algorithm. This algorithm compares profiles illustrating prediction dependencies on variable values generated by eXplainable Artificial Intelligence (XAI) techniques. To quantify differences in variable effects among models, we introduce the Profile Disparity Index (PDI) based on measures from functional data analysis. To illustrate the effectiveness of our approach, we showcase its application in predicting survival among hemophagocytic lymphohistiocytosis (HLH) patients - a foundational case study. Additionally, we benchmark our approach on other medical data sets, demonstrating its versatility and utility in various contexts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge