Evaluation of Machine Learning Methods to Predict Coronary Artery Disease Using Metabolomic Data
Paper and Code
Feb 28, 2017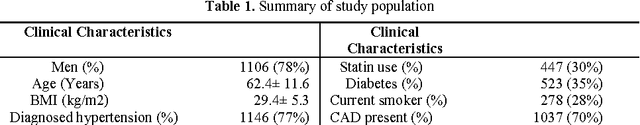
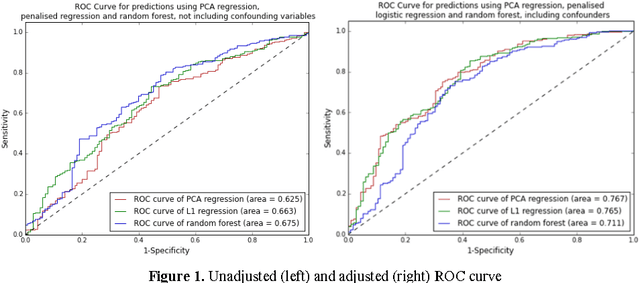
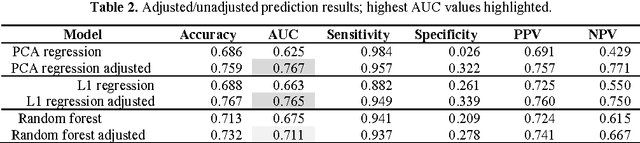
Metabolomic data can potentially enable accurate, non-invasive and low-cost prediction of coronary artery disease. Regression-based analytical approaches however might fail to fully account for interactions between metabolites, rely on a priori selected input features and thus might suffer from poorer accuracy. Supervised machine learning methods can potentially be used in order to fully exploit the dimensionality and richness of the data. In this paper, we systematically implement and evaluate a set of supervised learning methods (L1 regression, random forest classifier) and compare them to traditional regression-based approaches for disease prediction using metabolomic data.
* Medical Informatics Europe (MIE2017)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge