DNA data storage, sequencing data-carrying DNA
Paper and Code
May 11, 2022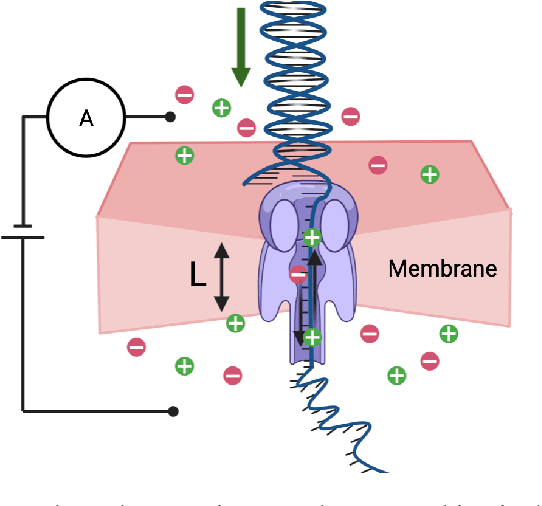
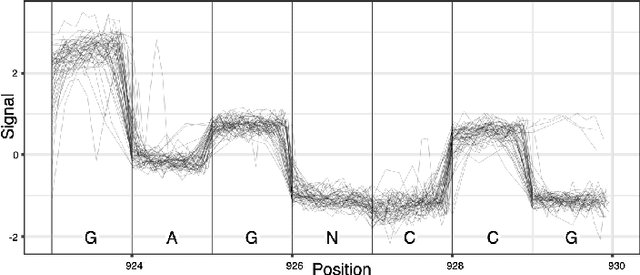
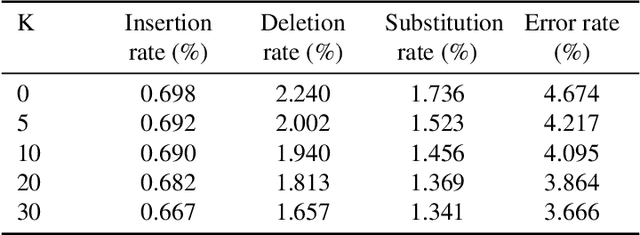
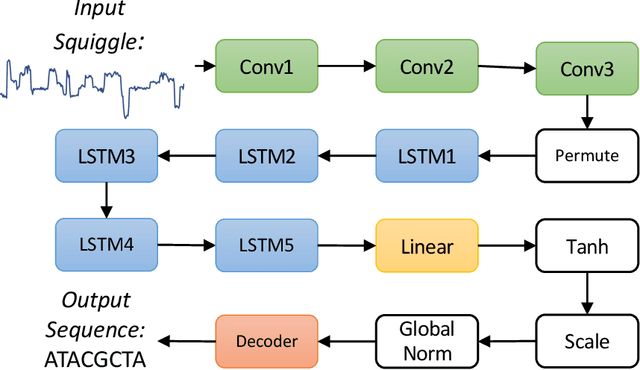
DNA is a leading candidate as the next archival storage media due to its density, durability and sustainability. To read (and write) data DNA storage exploits technology that has been developed over decades to sequence naturally occurring DNA in the life sciences. To achieve higher accuracy for previously unseen, biological DNA, sequencing relies on extending and training deep machine learning models known as basecallers. This growth in model complexity requires substantial resources, both computational and data sets. It also eliminates the possibility of a compact read head for DNA as a storage medium. We argue that we need to depart from blindly using sequencing models from the life sciences for DNA data storage. The difference is striking: for life science applications we have no control over the DNA, however, in the case of DNA data storage, we control how it is written, as well as the particular write head. More specifically, data-carrying DNA can be modulated and embedded with alignment markers and error correcting codes to guarantee higher fidelity and to carry out some of the work that the machine learning models perform. In this paper, we study accuracy trade-offs between deep model size and error correcting codes. We show that, starting with a model size of 107MB, the reduced accuracy from model compression can be compensated by using simple error correcting codes in the DNA sequences. In our experiments, we show that a substantial reduction in the size of the model does not incur an undue penalty for the error correcting codes used, therefore paving the way for portable data-carrying DNA read head. Crucially, we show that through the joint use of model compression and error correcting codes, we achieve a higher read accuracy than without compression and error correction codes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge