Distributional Principal Autoencoders
Paper and Code
Apr 21, 2024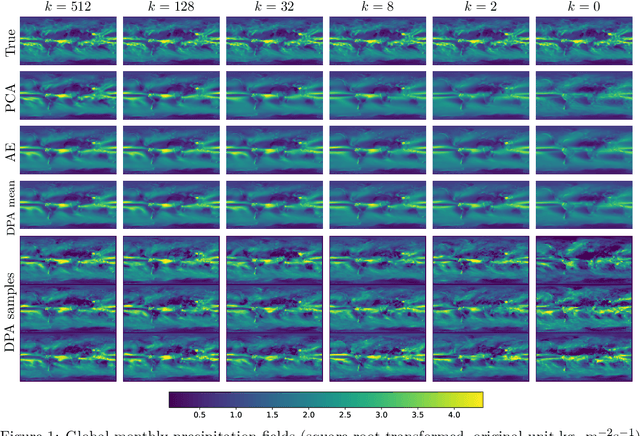
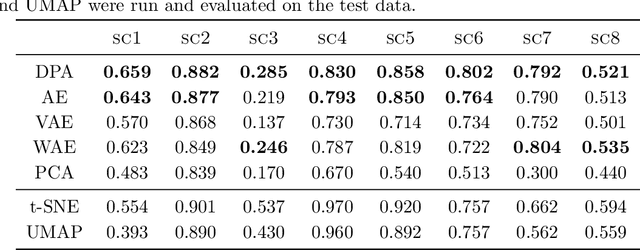
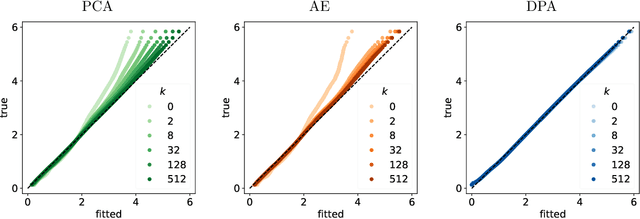
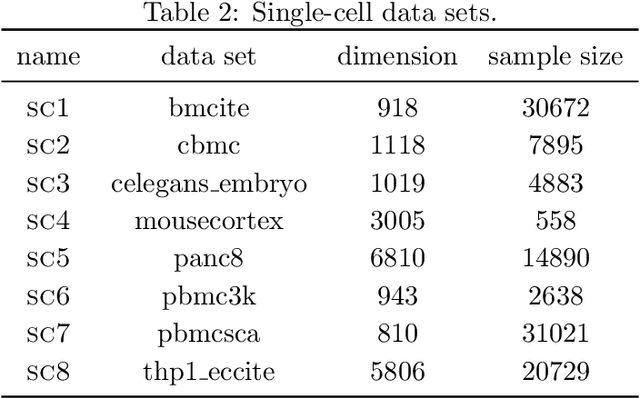
Dimension reduction techniques usually lose information in the sense that reconstructed data are not identical to the original data. However, we argue that it is possible to have reconstructed data identically distributed as the original data, irrespective of the retained dimension or the specific mapping. This can be achieved by learning a distributional model that matches the conditional distribution of data given its low-dimensional latent variables. Motivated by this, we propose Distributional Principal Autoencoder (DPA) that consists of an encoder that maps high-dimensional data to low-dimensional latent variables and a decoder that maps the latent variables back to the data space. For reducing the dimension, the DPA encoder aims to minimise the unexplained variability of the data with an adaptive choice of the latent dimension. For reconstructing data, the DPA decoder aims to match the conditional distribution of all data that are mapped to a certain latent value, thus ensuring that the reconstructed data retains the original data distribution. Our numerical results on climate data, single-cell data, and image benchmarks demonstrate the practical feasibility and success of the approach in reconstructing the original distribution of the data. DPA embeddings are shown to preserve meaningful structures of data such as the seasonal cycle for precipitations and cell types for gene expression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge