Direct Estimation of Difference Between Structural Equation Models in High Dimensions
Paper and Code
Jun 28, 2019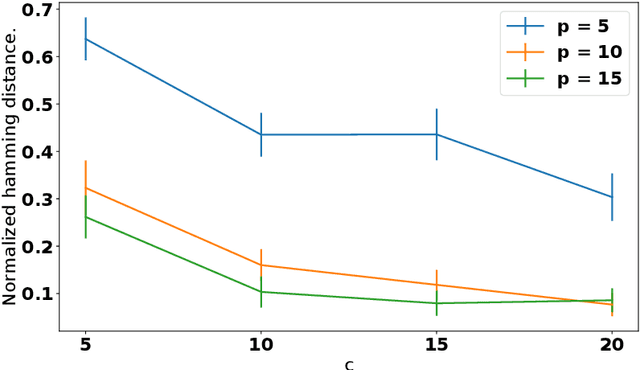
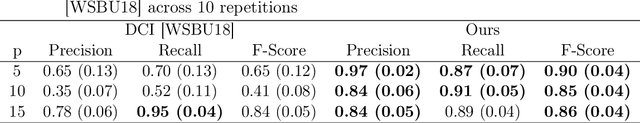
Discovering cause-effect relationships between variables from observational data is a fundamental challenge in many scientific disciplines. However, in many situations it is desirable to directly estimate the change in causal relationships across two different conditions, e.g., estimating the change in genetic expression across healthy and diseased subjects can help isolate genetic factors behind the disease. This paper focuses on the problem of directly estimating the structural difference between two causal DAGs, having the same topological ordering, given two sets of samples drawn from the individual DAGs. We present an algorithm that can recover the difference-DAG in $O(d \log p)$ samples, where $d$ is related to the number of edges in the difference-DAG. We also show that any method requires at least $\Omega(d \log p/d)$ samples to learn difference DAGs with at most $d$ parents per node. We validate our theoretical results with synthetic experiments and show that our method out-performs the state-of-the-art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge