Deep representation of EEG data from Spatio-Spectral Feature Images
Paper and Code
Jun 20, 2022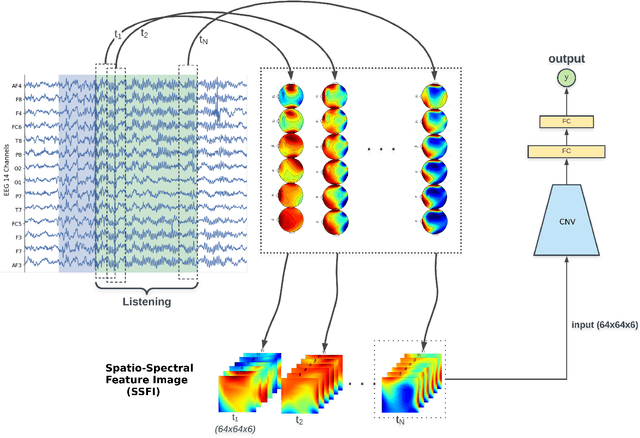
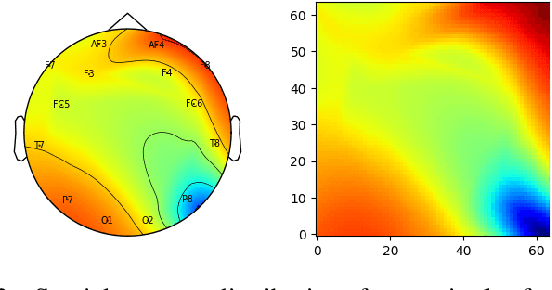
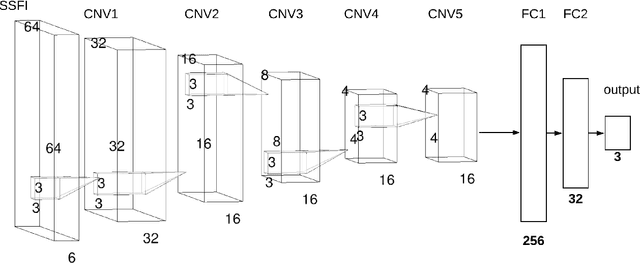
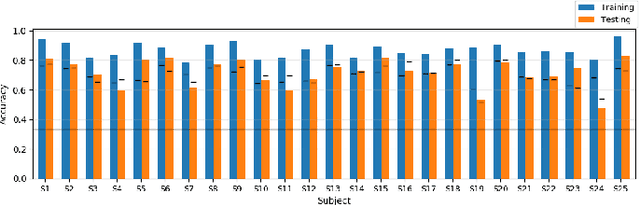
Unlike conventional data such as natural images, audio and speech, raw multi-channel Electroencephalogram (EEG) data are difficult to interpret. Modern deep neural networks have shown promising results in EEG studies, however finding robust invariant representations of EEG data across subjects remains a challenge, due to differences in brain folding structures. Thus, invariant representations of EEG data would be desirable to improve our understanding of the brain activity and to use them effectively during transfer learning. In this paper, we propose an approach to learn deep representations of EEG data by exploiting spatial relationships between recording electrodes and encoding them in a Spatio-Spectral Feature Images. We use multi-channel EEG signals from the PhyAAt dataset for auditory tasks and train a Convolutional Neural Network (CNN) on 25 subjects individually. Afterwards, we generate the input patterns that activate deep neurons across all the subjects. The generated pattern can be seen as a map of the brain activity in different spatial regions. Our analysis reveals the existence of specific brain regions related to different tasks. Low-level features focusing on larger regions and high-level features focusing on a smaller and very specific cluster of regions are also identified. Interestingly, similar patterns are found across different subjects although the activities appear in different regions. Our analysis also reveals common brain regions across subjects, which can be used as generalized representations. Our proposed approach allows us to find more interpretable representations of EEG data, which can further be used for effective transfer learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge