Deep Learning models for benign and malign Ocular Tumor Growth Estimation
Paper and Code
Jul 09, 2021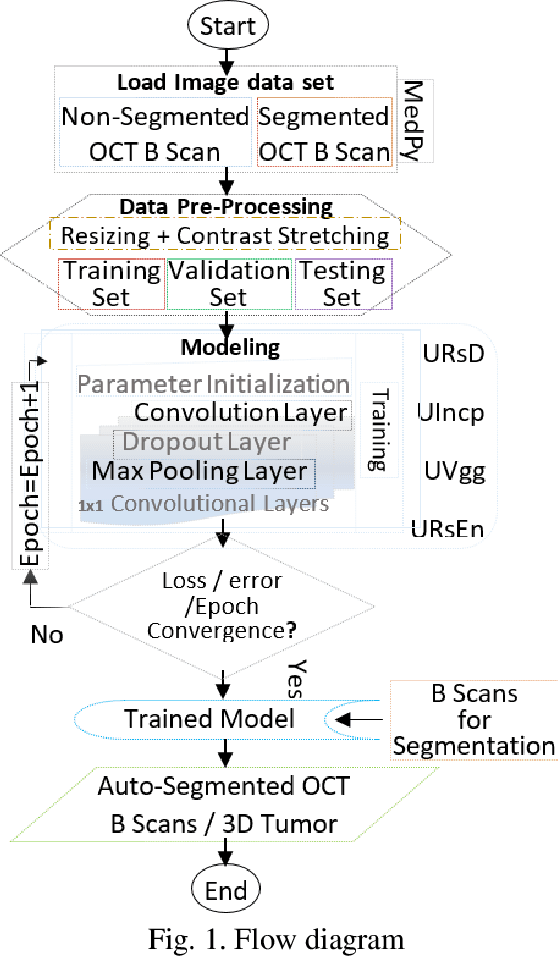


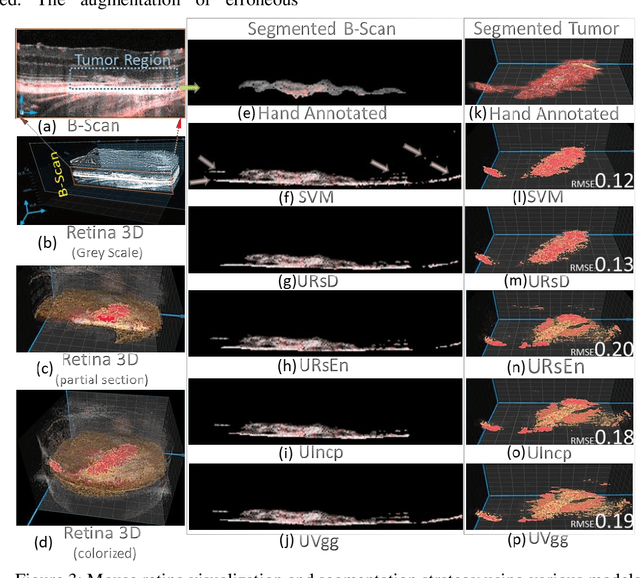
Relatively abundant availability of medical imaging data has provided significant support in the development and testing of Neural Network based image processing methods. Clinicians often face issues in selecting suitable image processing algorithm for medical imaging data. A strategy for the selection of a proper model is presented here. The training data set comprises optical coherence tomography (OCT) and angiography (OCT-A) images of 50 mice eyes with more than 100 days follow-up. The data contains images from treated and untreated mouse eyes. Four deep learning variants are tested for automatic (a) differentiation of tumor region with healthy retinal layer and (b) segmentation of 3D ocular tumor volumes. Exhaustive sensitivity analysis of deep learning models is performed with respect to the number of training and testing images using 8 eight performance indices to study accuracy, reliability/reproducibility, and speed. U-net with UVgg16 is best for malign tumor data set with treatment (having considerable variation) and U-net with Inception backbone for benign tumor data (with minor variation). Loss value and root mean square error (R.M.S.E.) are found most and least sensitive performance indices, respectively. The performance (via indices) is found to be exponentially improving regarding a number of training images. The segmented OCT-Angiography data shows that neovascularization drives the tumor volume. Image analysis shows that photodynamic imaging-assisted tumor treatment protocol is transforming an aggressively growing tumor into a cyst. An empirical expression is obtained to help medical professionals to choose a particular model given the number of images and types of characteristics. We recommend that the presented exercise should be taken as standard practice before employing a particular deep learning model for biomedical image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge