Continuous Representation of Molecules Using Graph Variational Autoencoder
Paper and Code
Apr 17, 2020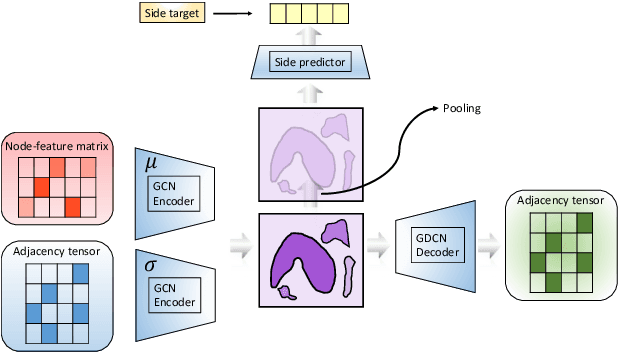
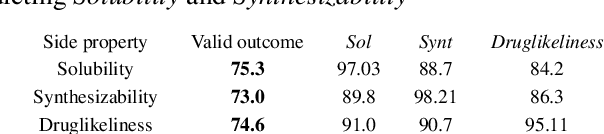
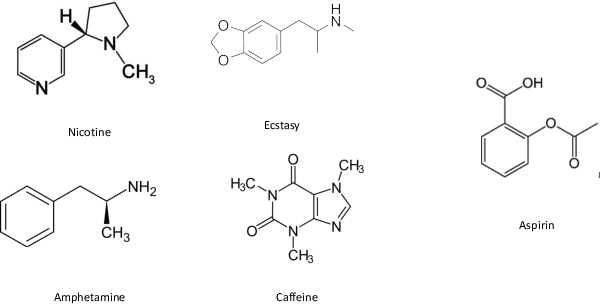
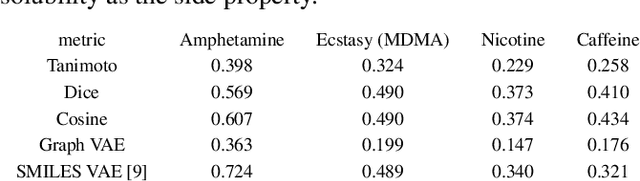
In order to continuously represent molecules, we propose a generative model in the form of a VAE which is operating on the 2D-graph structure of molecules. A side predictor is employed to prune the latent space and help the decoder in generating meaningful adjacency tensor of molecules. Other than the potential applicability in drug design and property prediction, we show the superior performance of this technique in comparison to other similar methods based on the SMILES representation of the molecules with RNN based encoder and decoder.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge