Computing the Hazard Ratios Associated with Explanatory Variables Using Machine Learning Models of Survival Data
Paper and Code
Feb 01, 2021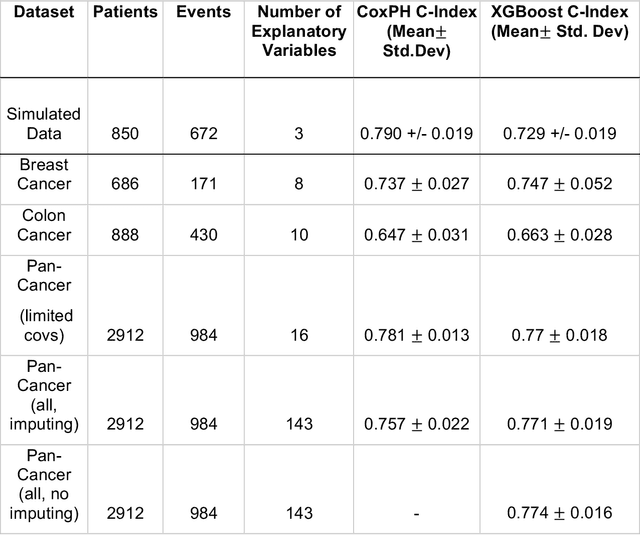

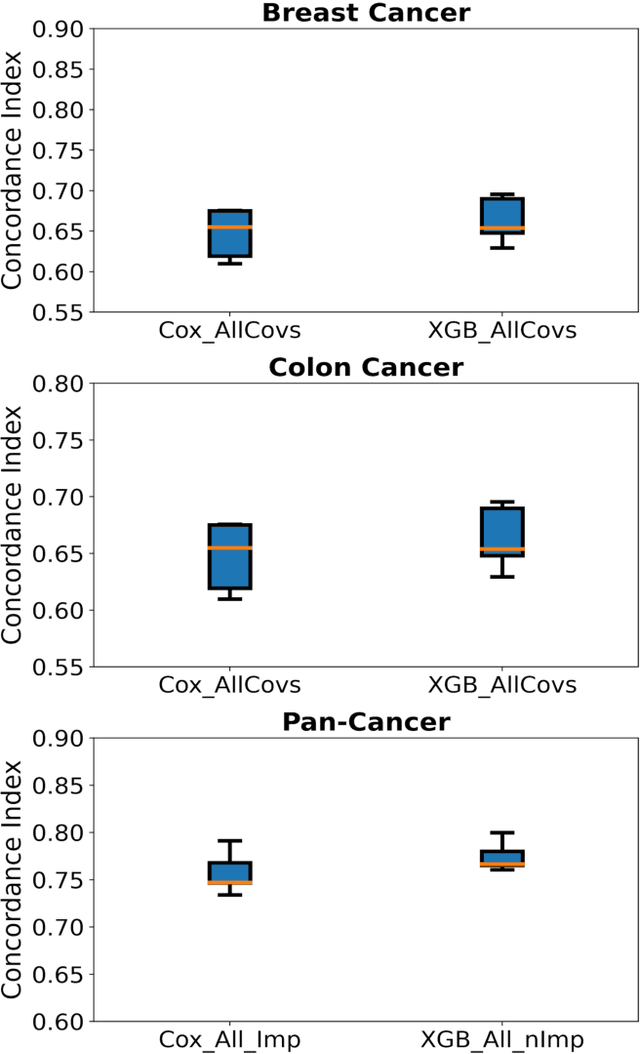
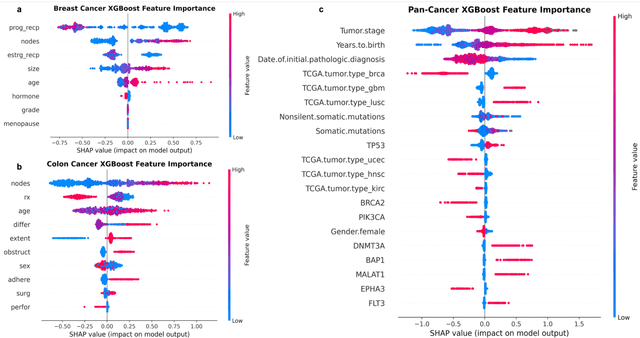
Purpose: The application of Cox Proportional Hazards (CoxPH) models to survival data and the derivation of Hazard Ratio (HR) is well established. While nonlinear, tree-based Machine Learning (ML) models have been developed and applied to the survival analysis, no methodology exists for computing HRs associated with explanatory variables from such models. We describe a novel way to compute HRs from tree-based ML models using the Shapley additive explanation (SHAP) values, which is a locally accurate and consistent methodology to quantify explanatory variables' contribution to predictions. Methods: We used three sets of publicly available survival data consisting of patients with colon, breast or pan cancer and compared the performance of CoxPH to the state-of-art ML model, XGBoost. To compute the HR for explanatory variables from the XGBoost model, the SHAP values were exponentiated and the ratio of the means over the two subgroups calculated. The confidence interval was computed via bootstrapping the training data and generating the ML model 1000 times. Across the three data sets, we systematically compared HRs for all explanatory variables. Open-source libraries in Python and R were used in the analyses. Results: For the colon and breast cancer data sets, the performance of CoxPH and XGBoost were comparable and we showed good consistency in the computed HRs. In the pan-cancer dataset, we showed agreement in most variables but also an opposite finding in two of the explanatory variables between the CoxPH and XGBoost result. Subsequent Kaplan-Meier plots supported the finding of the XGBoost model. Conclusion: Enabling the derivation of HR from ML models can help to improve the identification of risk factors from complex survival datasets and enhance the prediction of clinical trial outcomes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge