Clustering Gaussian Graphical Models
Paper and Code
Oct 05, 2019
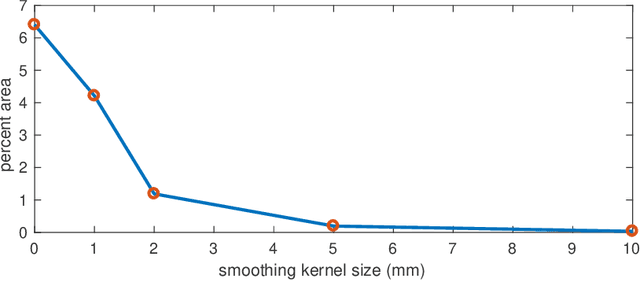
We derive an efficient method to perform clustering of nodes in Gaussian graphical models directly from sample data. Nodes are clustered based on the similarity of their network neighborhoods, with edge weights defined by partial correlations. In the limited-data scenario, where the covariance matrix would be rank-deficient, we are able to make use of matrix factors, and never need to estimate the actual covariance or precision matrix. We demonstrate the method on functional MRI data from the Human Connectome Project. A matlab implementation of the algorithm is provided.
* arXiv admin note: text overlap with arXiv:1903.07181
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge