Classification and Clustering of Sentence-Level Embeddings of Scientific Articles Generated by Contrastive Learning
Paper and Code
Mar 30, 2024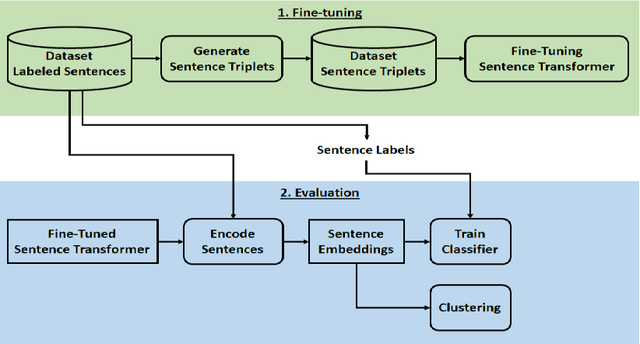


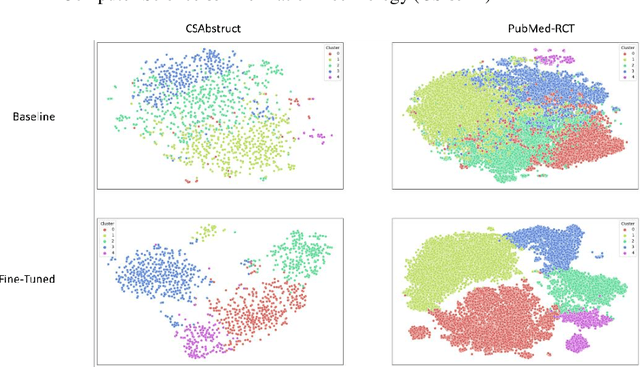
Scientific articles are long text documents organized into sections, each describing aspects of the research. Analyzing scientific production has become progressively challenging due to the increase in the number of available articles. Within this scenario, our approach consisted of fine-tuning transformer language models to generate sentence-level embeddings from scientific articles, considering the following labels: background, objective, methods, results, and conclusion. We trained our models on three datasets with contrastive learning. Two datasets are from the article's abstracts in the computer science and medical domains. Also, we introduce PMC-Sents-FULL, a novel dataset of sentences extracted from the full texts of medical articles. We compare the fine-tuned and baseline models in clustering and classification tasks to evaluate our approach. On average, clustering agreement measures values were five times higher. For the classification measures, in the best-case scenario, we had an average improvement in F1-micro of 30.73\%. Results show that fine-tuning sentence transformers with contrastive learning and using the generated embeddings in downstream tasks is a feasible approach to sentence classification in scientific articles. Our experiment codes are available on GitHub.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge