Class-Conditional VAE-GAN for Local-Ancestry Simulation
Paper and Code
Nov 27, 2019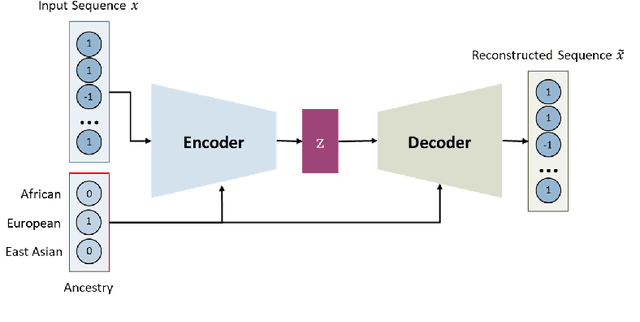
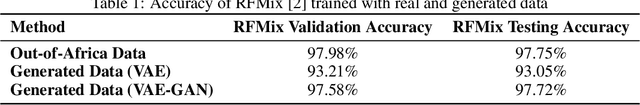
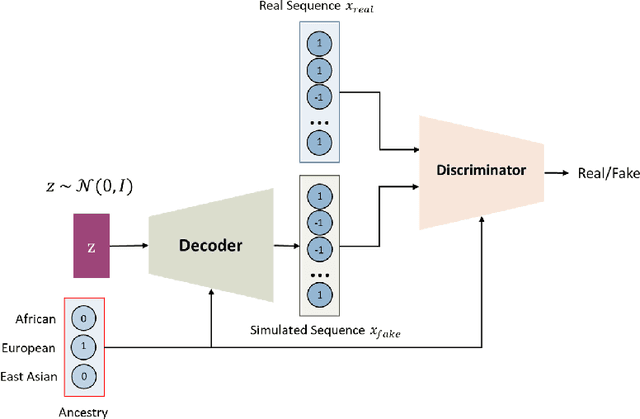
Local ancestry inference (LAI) allows identification of the ancestry of all chromosomal segments in admixed individuals, and it is a critical step in the analysis of human genomes with applications from pharmacogenomics and precision medicine to genome-wide association studies. In recent years, many LAI techniques have been developed in both industry and academic research. However, these methods require large training data sets of human genomic sequences from the ancestries of interest. Such reference data sets are usually limited, proprietary, protected by privacy restrictions, or otherwise not accessible to the public. Techniques to generate training samples that resemble real haploid sequences from ancestries of interest can be useful tools in such scenarios, since a generalized model can often be shared, but the unique human sample sequences cannot. In this work we present a class-conditional VAE-GAN to generate new human genomic sequences that can be used to train local ancestry inference (LAI) algorithms. We evaluate the quality of our generated data by comparing the performance of a state-of-the-art LAI method when trained with generated versus real data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge