CHERRY: a Computational metHod for accuratE pRediction of virus-pRokarYotic interactions using a graph encoder-decoder model
Paper and Code
Jan 04, 2022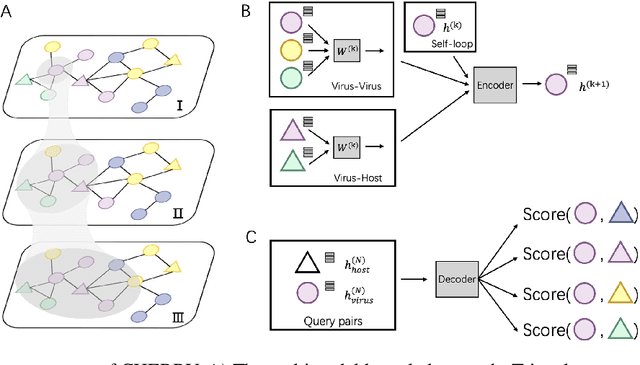
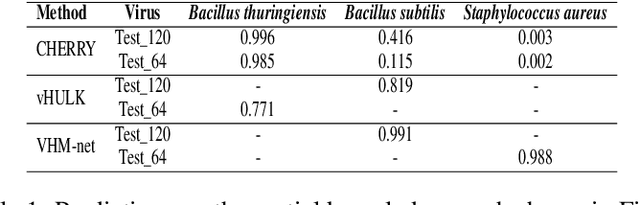
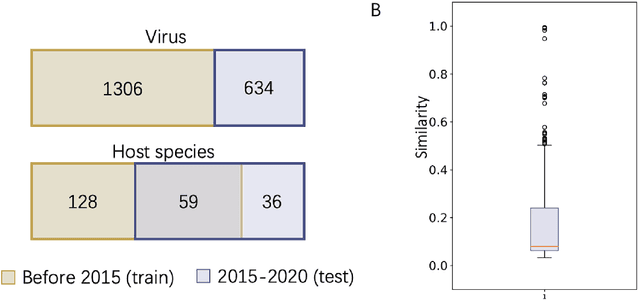
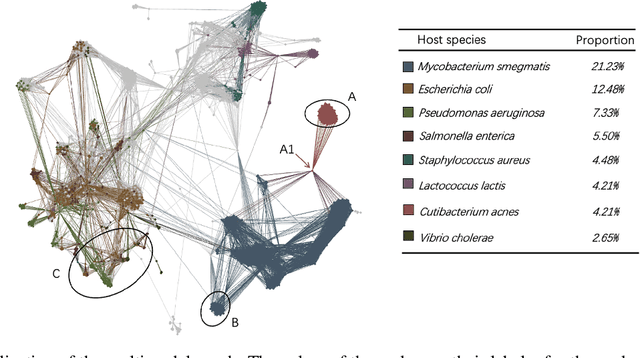
Prokaryotic viruses, which infect bacteria and archaea, are key players in microbial communities. Predicting the hosts of prokaryotic viruses helps decipher the dynamic relationship between microbes. Although there are experimental methods for host identification, they are either labor-intensive or require the cultivation of the host cells, creating a need for computational host prediction. Despite some promising results, computational host prediction remains a challenge because of the limited known interactions and the sheer amount of sequenced phages by high-throughput sequencing technologies. The state-of-the-art methods can only achieve 43% accuracy at the species level. This work presents CHERRY, a tool formulating host prediction as link prediction in a knowledge graph. As a virus-prokaryotic interaction prediction tool, CHERRY can be applied to predict hosts for newly discovered viruses and also the viruses infecting antibiotic-resistant bacteria. We demonstrated the utility of CHERRY for both applications and compared its performance with the state-of-the-art methods in different scenarios. To our best knowledge, CHERRY has the highest accuracy in identifying virus-prokaryote interactions. It outperforms all the existing methods at the species level with an accuracy increase of 37%. In addition, CHERRY's performance is more stable on short contigs than other tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge