CD&S Dataset: Handheld Imagery Dataset Acquired Under Field Conditions for Corn Disease Identification and Severity Estimation
Paper and Code
Oct 22, 2021
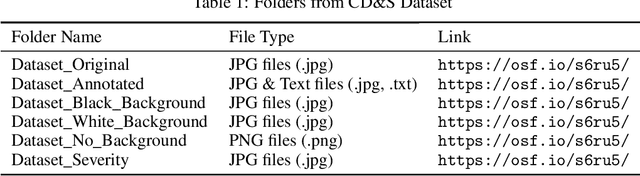
Accurate disease identification and its severity estimation is an important consideration for disease management. Deep learning-based solutions for disease management using imagery datasets are being increasingly explored by the research community. However, most reported studies have relied on imagery datasets that were acquired under controlled lab conditions. As a result, such models lacked the ability to identify diseases in the field. Therefore, to train a robust deep learning model for field use, an imagery dataset was created using raw images acquired under field conditions using a handheld sensor and augmented images with varying backgrounds. The Corn Disease and Severity (CD&S) dataset consisted of 511, 524, and 562, field acquired raw images, corresponding to three common foliar corn diseases, namely Northern Leaf Blight (NLB), Gray Leaf Spot (GLS), and Northern Leaf Spot (NLS), respectively. For training disease identification models, half of the imagery data for each disease was annotated using bounding boxes and also used to generate 2343 additional images through augmentation using three different backgrounds. For severity estimation, an additional 515 raw images for NLS were acquired and categorized into severity classes ranging from 1 (resistant) to 5 (susceptible). Overall, the CD&S dataset consisted of 4455 total images comprising of 2112 field images and 2343 augmented images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge