Canonical Polyadic Decomposition with Auxiliary Information for Brain Computer Interface
Paper and Code
Nov 05, 2015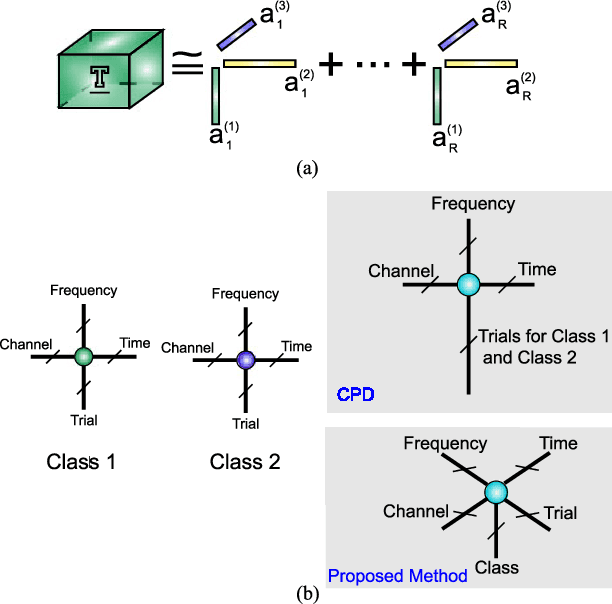
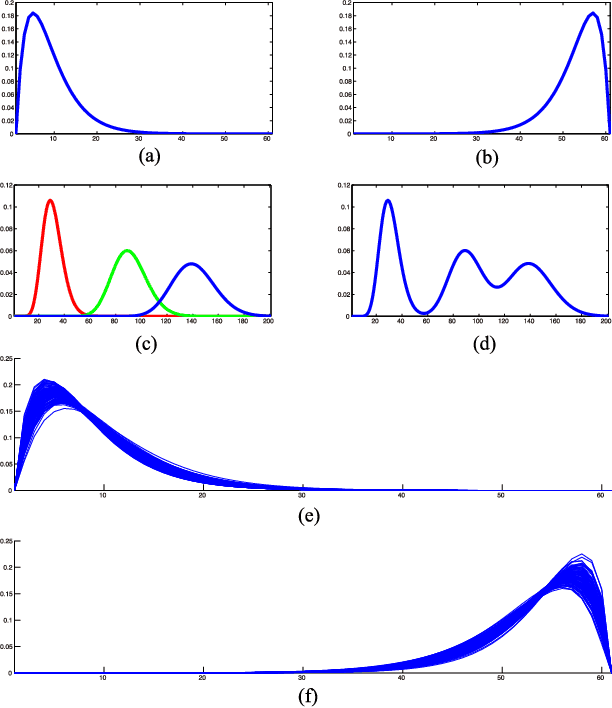
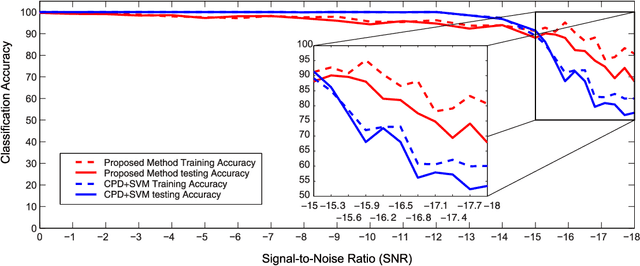
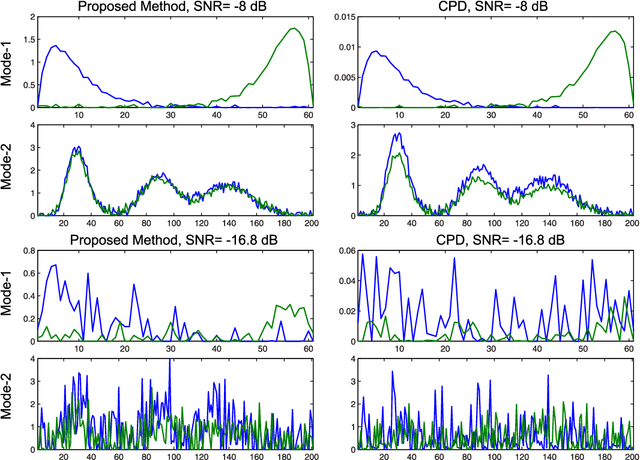
Physiological signals are often organized in the form of multiple dimensions (e.g., channel, time, task, and 3D voxel), so it is better to preserve original organization structure when processing. Unlike vector-based methods that destroy data structure, Canonical Polyadic Decomposition (CPD) aims to process physiological signals in the form of multi-way array, which considers relationships between dimensions and preserves structure information contained by the physiological signal. Nowadays, CPD is utilized as an unsupervised method for feature extraction in a classification problem. After that, a classifier, such as support vector machine, is required to classify those features. In this manner, classification task is achieved in two isolated steps. We proposed supervised Canonical Polyadic Decomposition by directly incorporating auxiliary label information during decomposition, by which a classification task can be achieved without an extra step of classifier training. The proposed method merges the decomposition and classifier learning together, so it reduces procedure of classification task compared with that of respective decomposition and classification. In order to evaluate the performance of the proposed method, three different kinds of signals, synthetic signal, EEG signal, and MEG signal, were used. The results based on evaluations of synthetic and real signals demonstrated that the proposed method is effective and efficient.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge