Benchmarking BioRelEx for Entity Tagging and Relation Extraction
Paper and Code
May 31, 2020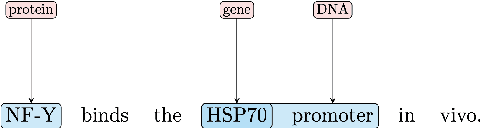
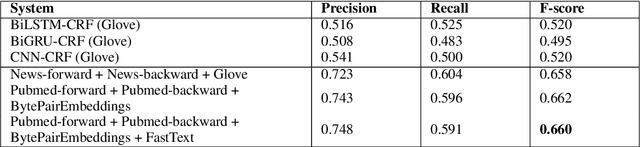
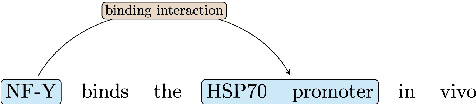
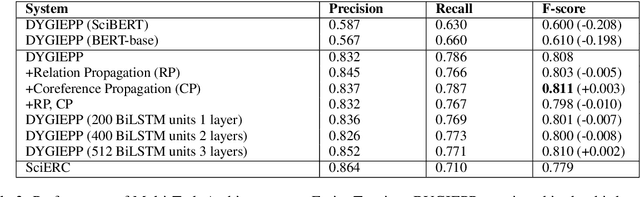
Extracting relationships and interactions between different biological entities is still an extremely challenging problem but has not received much attention as much as extraction in other generic domains. In addition to the lack of annotated data, low benchmarking is still a major reason for slow progress. In order to fill this gap, we compare multiple existing entity and relation extraction models over a recently introduced public dataset, BioRelEx of sentences annotated with biological entities and relations. Our straightforward benchmarking shows that span-based multi-task architectures like DYGIE show 4.9% and 6% absolute improvements in entity tagging and relation extraction respectively over the previous state-of-art and that incorporating domain-specific information like embeddings pre-trained over related domains boosts performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge