Atomistic structure search using local surrogate mode
Paper and Code
Aug 19, 2022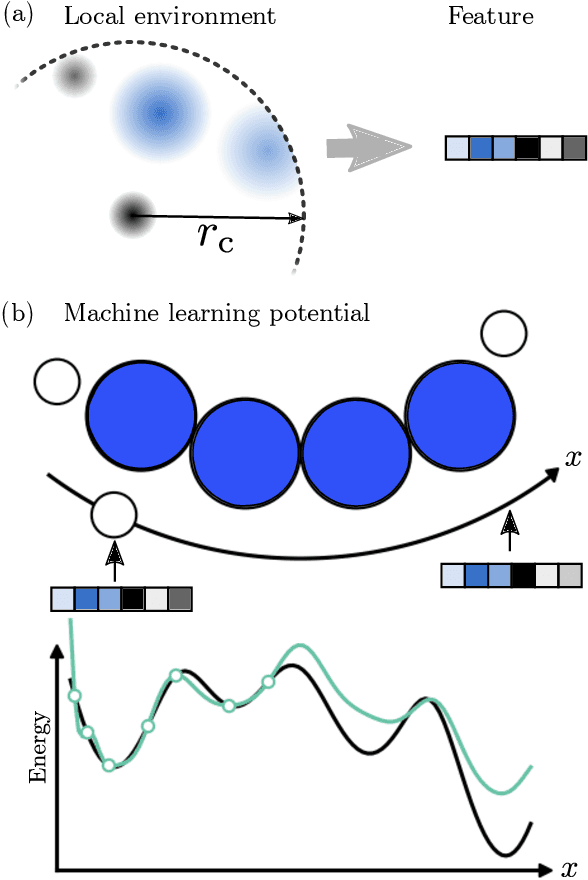
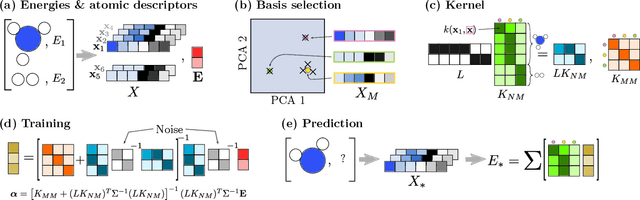
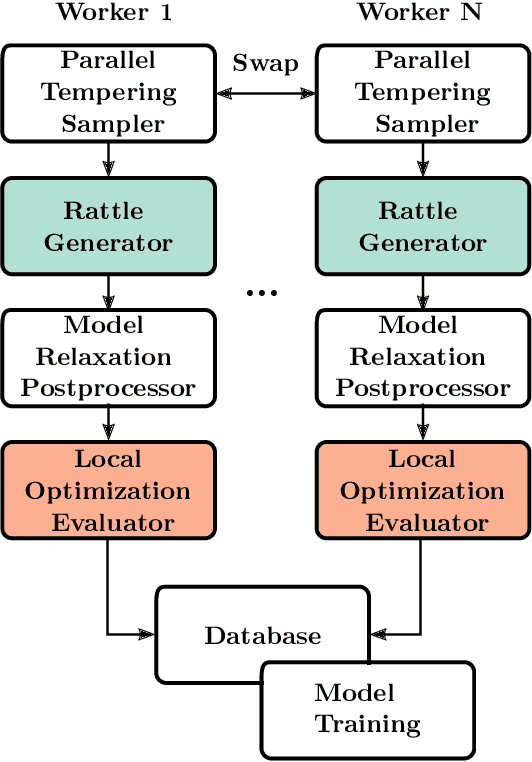

We describe a local surrogate model for use in conjunction with global structure search methods. The model follows the Gaussian approximation potential (GAP) formalism and is based on a the smooth overlap of atomic positions descriptor with sparsification in terms of a reduced number of local environments using mini-batch $k$-means. The model is implemented in the Atomistic Global Optimization X framework and used as a partial replacement of the local relaxations in basin hopping structure search. The approach is shown to be robust for a wide range of atomistic system including molecules, nano-particles, surface supported clusters and surface thin films. The benefits in a structure search context of a local surrogate model are demonstrated. This includes the ability to transfer learning from smaller systems as well as the possibility to perform concurrent multi-stoichiometry searches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge