An iterative feature selection method for GRNs inference by exploring topological properties
Paper and Code
Jul 25, 2011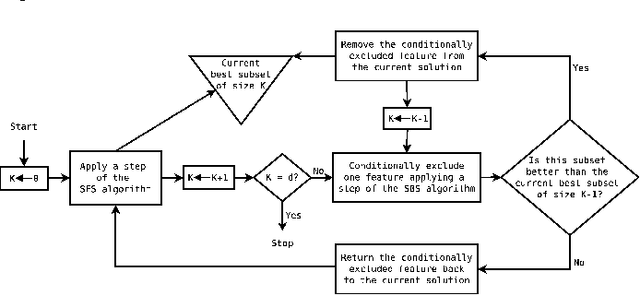
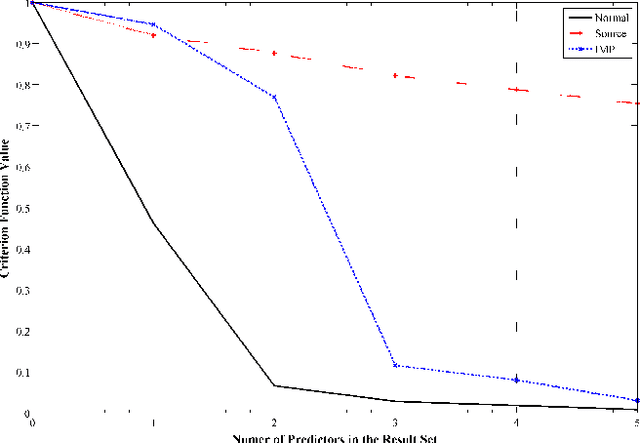
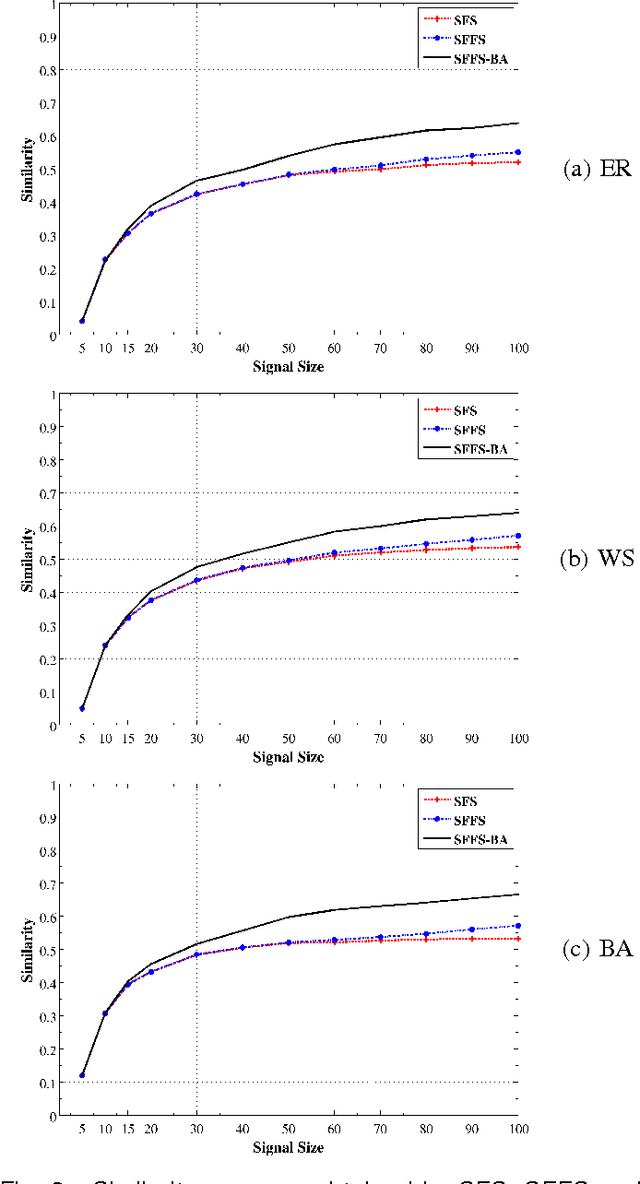
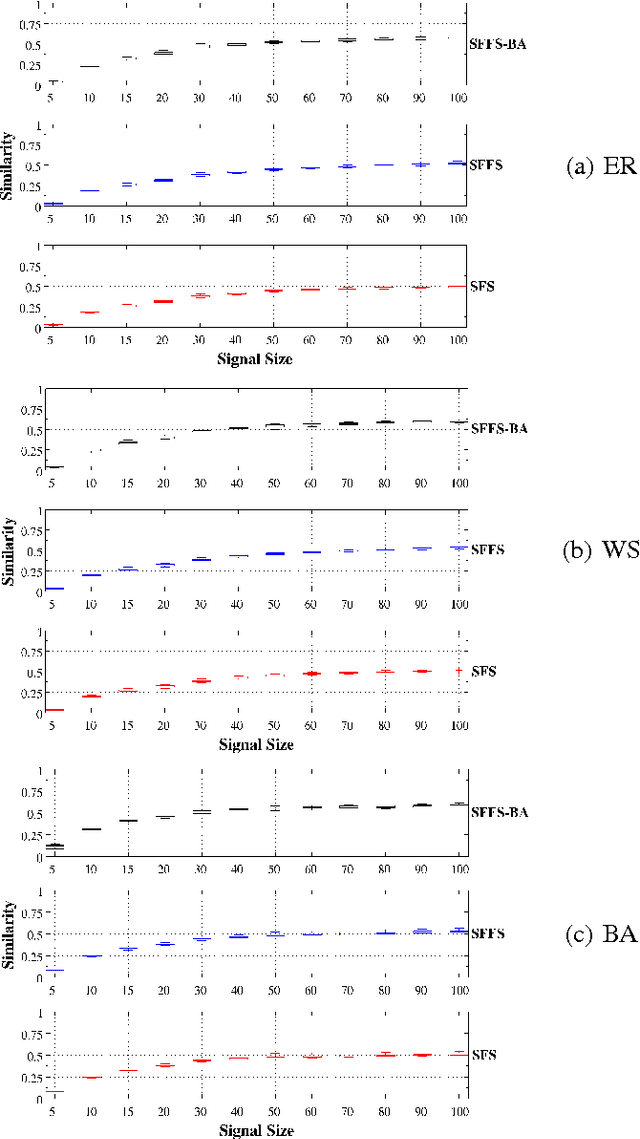
An important problem in bioinformatics is the inference of gene regulatory networks (GRN) from temporal expression profiles. In general, the main limitations faced by GRN inference methods is the small number of samples with huge dimensionalities and the noisy nature of the expression measurements. In face of these limitations, alternatives are needed to get better accuracy on the GRNs inference problem. This work addresses this problem by presenting an alternative feature selection method that applies prior knowledge on its search strategy, called SFFS-BA. The proposed search strategy is based on the Sequential Floating Forward Selection (SFFS) algorithm, with the inclusion of a scale-free (Barab\'asi-Albert) topology information in order to guide the search process to improve inference. The proposed algorithm explores the scale-free property by pruning the search space and using a power law as a weight for reducing it. In this way, the search space traversed by the SFFS-BA method combines a breadth-first search when the number of combinations is small (<k> <= 2) with a depth-first search when the number of combinations becomes explosive (<k> >= 3), being guided by the scale-free prior information. Experimental results show that the SFFS-BA provides a better inference similarities than SFS and SFFS, keeping the robustness of the SFS and SFFS methods, thus presenting very good results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge