A Supervised Machine Learning Approach for Sequence Based Protein-protein Interaction (PPI) Prediction
Paper and Code
Mar 27, 2022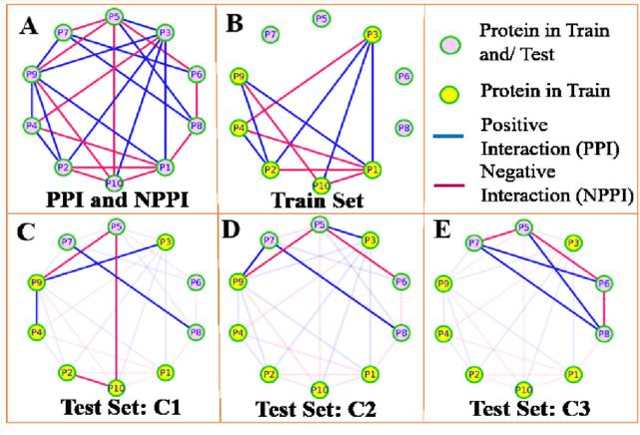

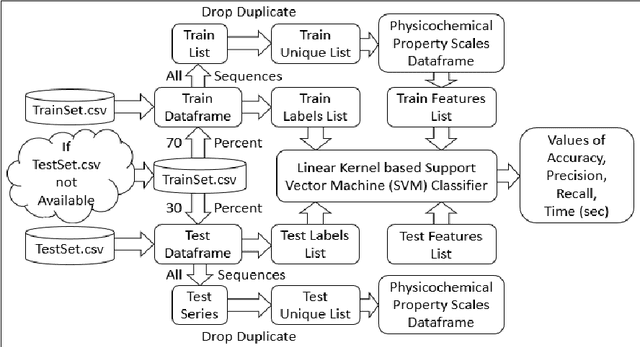

Computational protein-protein interaction (PPI) prediction techniques can contribute greatly in reducing time, cost and false-positive interactions compared to experimental approaches. Sequence is one of the key and primary information of proteins that plays a crucial role in PPI prediction. Several machine learning approaches have been applied to exploit the characteristics of PPI datasets. However, these datasets greatly influence the performance of predicting models. So, care should be taken on both dataset curation as well as design of predictive models. Here, we have described our submitted solution with the results of the SeqPIP competition whose objective was to develop comprehensive PPI predictive models from sequence information with high-quality bias-free interaction datasets. A training set of 2000 positive and 2000 negative interactions with sequences was given to us. Our method was evaluated with three independent high-quality interaction test datasets and with other competitors solutions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge