A scalable approach for developing clinical risk prediction applications in different hospitals
Paper and Code
Jan 21, 2021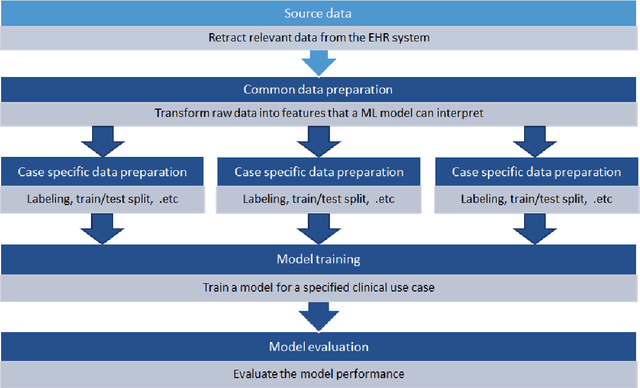
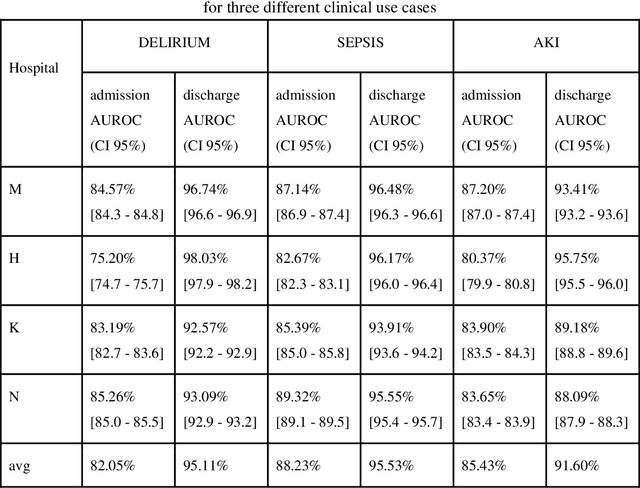

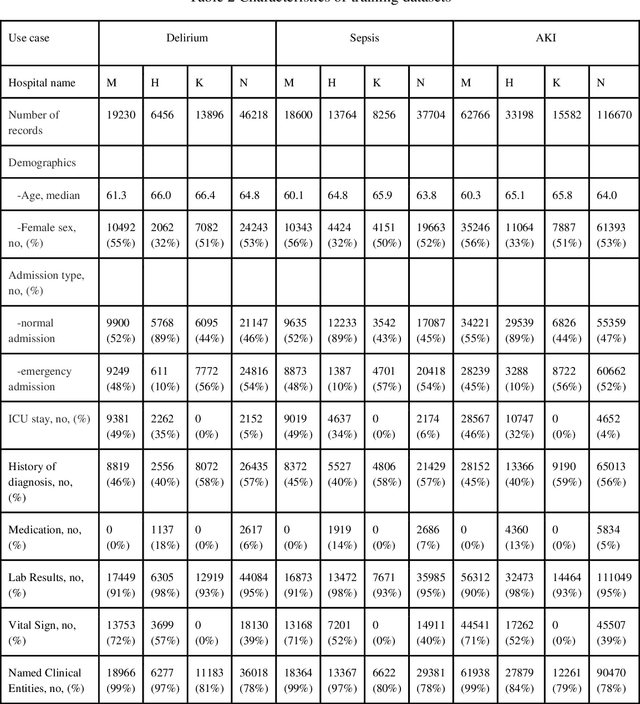
Objective: Machine learning algorithms are now widely used in predicting acute events for clinical applications. While most of such prediction applications are developed to predict the risk of a particular acute event at one hospital, few efforts have been made in extending the developed solutions to other events or to different hospitals. We provide a scalable solution to extend the process of clinical risk prediction model development of multiple diseases and their deployment in different Electronic Health Records (EHR) systems. Materials and Methods: We defined a generic process for clinical risk prediction model development. A calibration tool has been created to automate the model generation process. We applied the model calibration process at four hospitals, and generated risk prediction models for delirium, sepsis and acute kidney injury (AKI) respectively at each of these hospitals. Results: The delirium risk prediction models achieved area under the receiver-operating characteristic curve (AUROC) ranging from 0.82 to 0.95 over different stages of a hospital stay on the test datasets of the four hospitals. The sepsis models achieved AUROC ranging from 0.88 to 0.95, and the AKI models achieved AUROC ranging from 0.85 to 0.92. Discussion: The scalability discussed in this paper is based on building common data representations (syntactic interoperability) between EHRs stored in different hospitals. Semantic interoperability, a more challenging requirement that different EHRs share the same meaning of data, e.g. a same lab coding system, is not mandated with our approach. Conclusions: Our study describes a method to develop and deploy clinical risk prediction models in a scalable way. We demonstrate its feasibility by developing risk prediction models for three diseases across four hospitals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge