Zhongqiang Liu
A comparative study on machine learning approaches for rock mass classification using drilling data
Mar 15, 2024



Abstract:Current rock engineering design in drill and blast tunnelling primarily relies on engineers' observational assessments. Measure While Drilling (MWD) data, a high-resolution sensor dataset collected during tunnel excavation, is underutilised, mainly serving for geological visualisation. This study aims to automate the translation of MWD data into actionable metrics for rock engineering. It seeks to link data to specific engineering actions, thus providing critical decision support for geological challenges ahead of the tunnel face. Leveraging a large and geologically diverse dataset of 500,000 drillholes from 15 tunnels, the research introduces models for accurate rock mass quality classification in a real-world tunnelling context. Both conventional machine learning and image-based deep learning are explored to classify MWD data into Q-classes and Q-values, examples of metrics describing the stability of the rock mass, using both tabular and image data. The results indicate that the K-nearest neighbours algorithm in an ensemble with tree-based models using tabular data, effectively classifies rock mass quality. It achieves a cross-validated balanced accuracy of 0.86 in classifying rock mass into the Q-classes A, B, C, D, E1, E2, and 0.95 for a binary classification with E versus the rest. Classification using a CNN with MWD-images for each blasting round resulted in a balanced accuracy of 0.82 for binary classification. Regressing the Q-value from tabular MWD-data achieved cross-validated R2 and MSE scores of 0.80 and 0.18 for a similar ensemble model as in classification. High performance in regression and classification boosts confidence in automated rock mass assessment. Applying advanced modelling on a unique dataset demonstrates MWD data's value in improving rock mass classification accuracy and advancing data-driven rock engineering design, reducing manual intervention.
Automatic Segmentation of Non-Tumor Tissues in Glioma MR Brain Images Using Deformable Registration with Partial Convolutional Networks
Jul 10, 2020

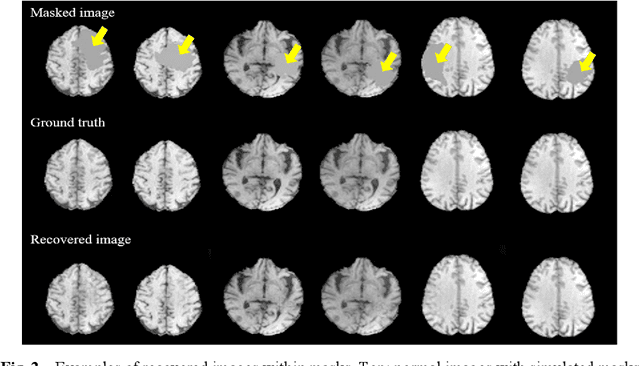
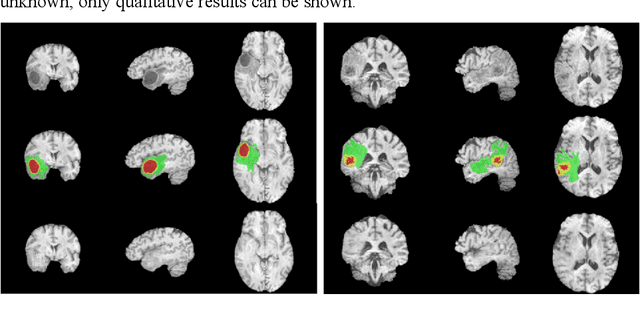
Abstract:In brain tumor diagnosis and surgical planning, segmentation of tumor regions and accurate analysis of surrounding normal tissues are necessary for physicians. Pathological variability often renders difficulty to register a well-labeled normal atlas to such images and to automatic segment/label surrounding normal brain tissues. In this paper, we propose a new registration approach that first segments brain tumor using a U-Net and then simulates missed normal tissues within the tumor region using a partial convolutional network. Then, a standard normal brain atlas image is registered onto such tumor-removed images in order to segment/label the normal brain tissues. In this way, our new approach greatly reduces the effects of pathological variability in deformable registration and segments the normal tissues surrounding brain tumor well. In experiments, we used MICCAI BraTS2018 T1 tumor images to evaluate the proposed algorithm. By comparing direct registration with the proposed algorithm, the results showed that the Dice coefficient for gray matters was significantly improved for surrounding normal brain tissues.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge