Tong-Tian Weng
CAS Key Laboratory of Quantum Information, University of Science and Technology of China, Hefei, 230026, China, CAS Center For Excellence in Quantum Information and Quantum Physics, University of Science and Technology of China, Hefei, 230026, China
Enhancing Deep Learning Based Structured Illumination Microscopy Reconstruction with Light Field Awareness
Mar 14, 2025Abstract:Structured illumination microscopy (SIM) is a pivotal technique for dynamic subcellular imaging in live cells. Conventional SIM reconstruction algorithms depend on accurately estimating the illumination pattern and can introduce artefacts when this estimation is imprecise. Although recent deep learning-based SIM reconstruction methods have improved speed, accuracy, and robustness, they often struggle with out-of-distribution data. To address this limitation, we propose an Awareness-of-Light-field SIM (AL-SIM) reconstruction approach that directly estimates the actual light field to correct for errors arising from data distribution shifts. Through comprehensive experiments on both simulated filament structures and live BSC1 cells, our method demonstrates a 7% reduction in the normalized root mean square error (NRMSE) and substantially lowers reconstruction artefacts. By minimizing these artefacts and improving overall accuracy, AL-SIM broadens the applicability of SIM for complex biological systems.
Learning imaging mechanism directly from optical microscopy observations
Apr 25, 2023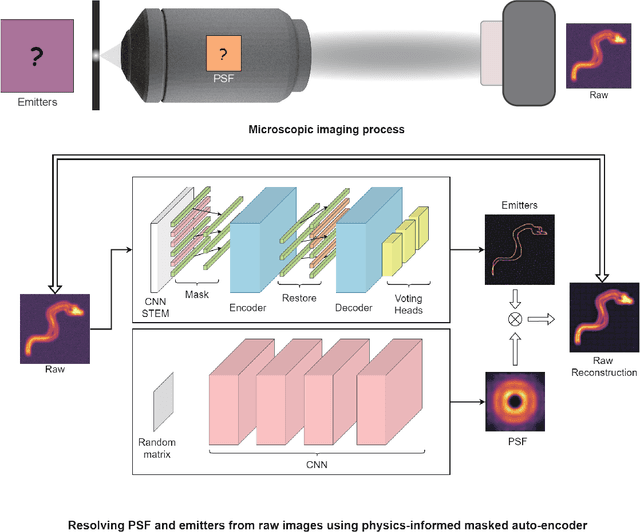
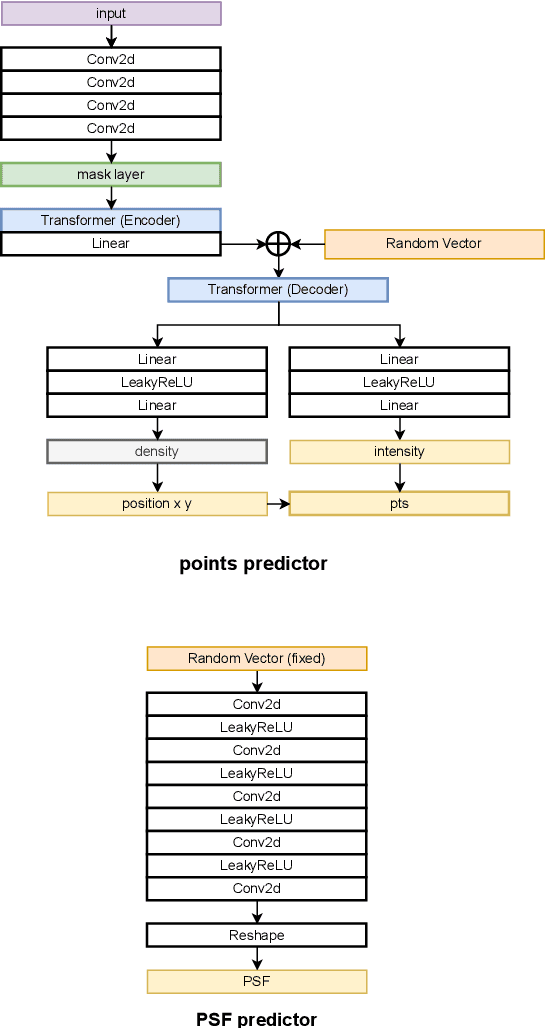
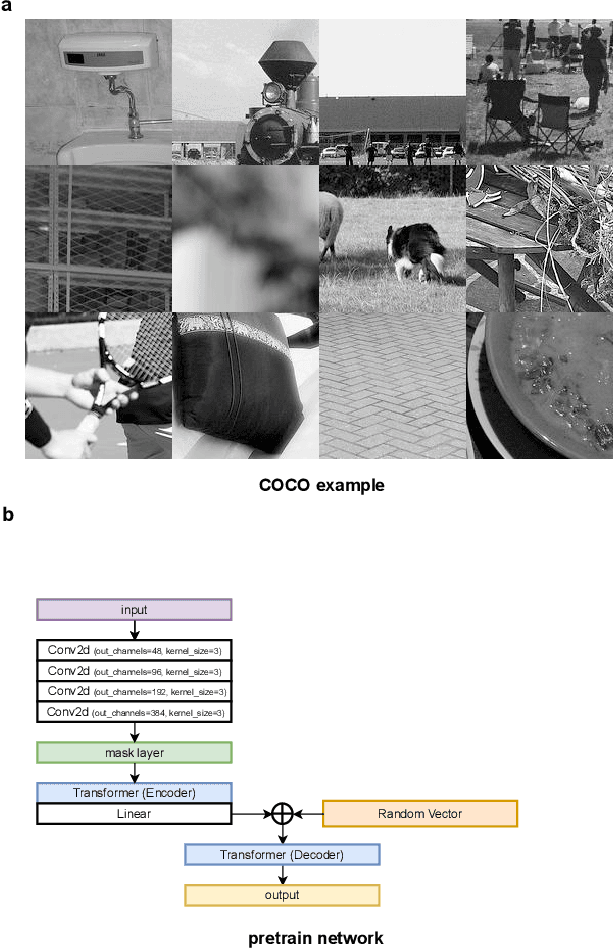

Abstract:Optical microscopy image plays an important role in scientific research through the direct visualization of the nanoworld, where the imaging mechanism is described as the convolution of the point spread function (PSF) and emitters. Based on a priori knowledge of the PSF or equivalent PSF, it is possible to achieve more precise exploration of the nanoworld. However, it is an outstanding challenge to directly extract the PSF from microscopy images. Here, with the help of self-supervised learning, we propose a physics-informed masked autoencoder (PiMAE) that enables a learnable estimation of the PSF and emitters directly from the raw microscopy images. We demonstrate our method in synthetic data and real-world experiments with significant accuracy and noise robustness. PiMAE outperforms DeepSTORM and the Richardson-Lucy algorithm in synthetic data tasks with an average improvement of 19.6\% and 50.7\% (35 tasks), respectively, as measured by the normalized root mean square error (NRMSE) metric. This is achieved without prior knowledge of the PSF, in contrast to the supervised approach used by DeepSTORM and the known PSF assumption in the Richardson-Lucy algorithm. Our method, PiMAE, provides a feasible scheme for achieving the hidden imaging mechanism in optical microscopy and has the potential to learn hidden mechanisms in many more systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge