Simon Wein
Data Integration with Fusion Searchlight: Classifying Brain States from Resting-state fMRI
Dec 13, 2024



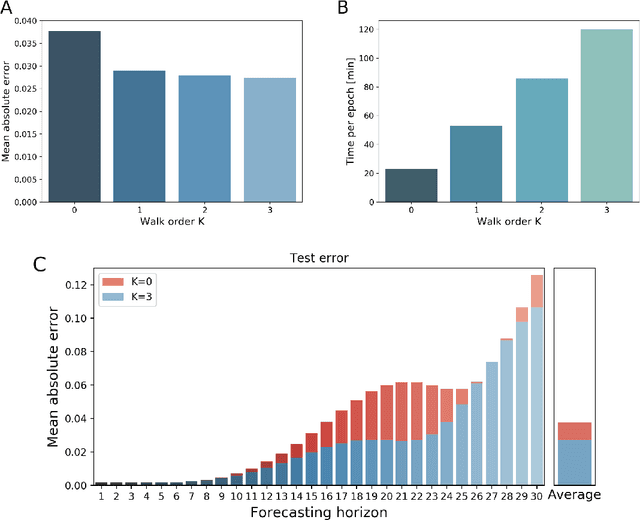
Abstract:Spontaneous neural activity observed in resting-state fMRI is characterized by complex spatio-temporal dynamics. Different measures related to local and global brain connectivity and fluctuations in low-frequency amplitudes can quantify individual aspects of these neural dynamics. Even though such measures are derived from the same functional signals, they are often evaluated separately, neglecting their interrelations and potentially reducing the analysis sensitivity. In our study, we present a fusion searchlight (FuSL) framework to combine the complementary information contained in different resting-state fMRI metrics and demonstrate how this can improve the decoding of brain states. Moreover, we show how explainable AI allows us to reconstruct the differential impact of each metric on the decoding, which additionally increases spatial specificity of searchlight analysis. In general, this framework can be adapted to combine information derived from different imaging modalities or experimental conditions, offering a versatile and interpretable tool for data fusion in neuroimaging.
Modeling Spatio-Temporal Dynamics in Brain Networks: A Comparison of Graph Neural Network Architectures
Dec 08, 2021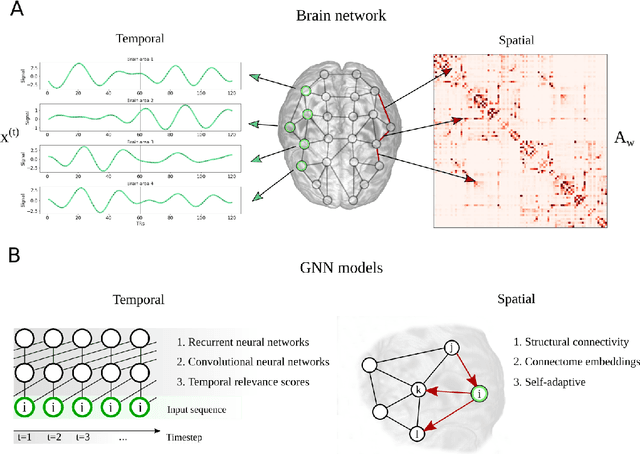

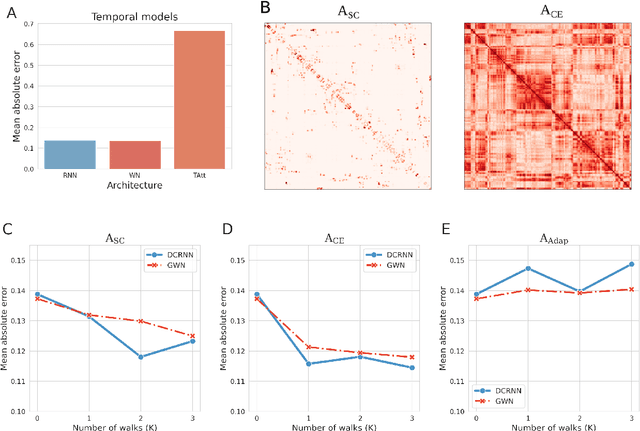
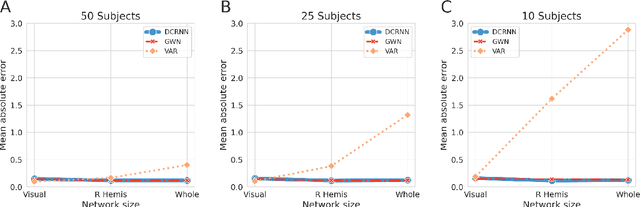
Abstract:Comprehending the interplay between spatial and temporal characteristics of neural dynamics can contribute to our understanding of information processing in the human brain. Graph neural networks (GNNs) provide a new possibility to interpret graph structured signals like those observed in complex brain networks. In our study we compare different spatio-temporal GNN architectures and study their ability to replicate neural activity distributions obtained in functional MRI (fMRI) studies. We evaluate the performance of the GNN models on a variety of scenarios in MRI studies and also compare it to a VAR model, which is currently predominantly used for directed functional connectivity analysis. We show that by learning localized functional interactions on the anatomical substrate, GNN based approaches are able to robustly scale to large network studies, even when available data are scarce. By including anatomical connectivity as the physical substrate for information propagation, such GNNs also provide a multimodal perspective on directed connectivity analysis, offering a novel possibility to investigate the spatio-temporal dynamics in brain networks.
A Graph Neural Network Framework for Causal Inference in Brain Networks
Oct 14, 2020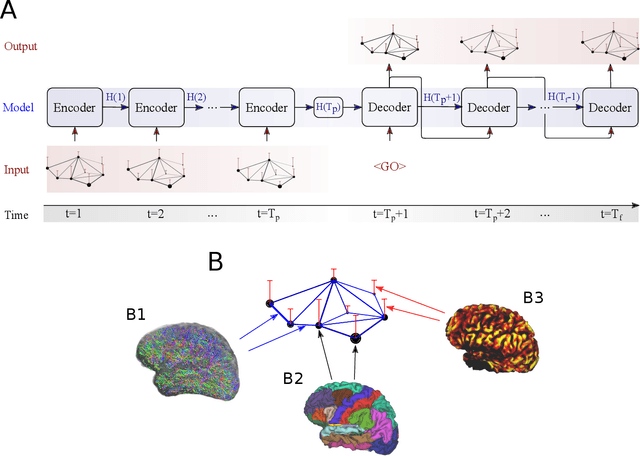

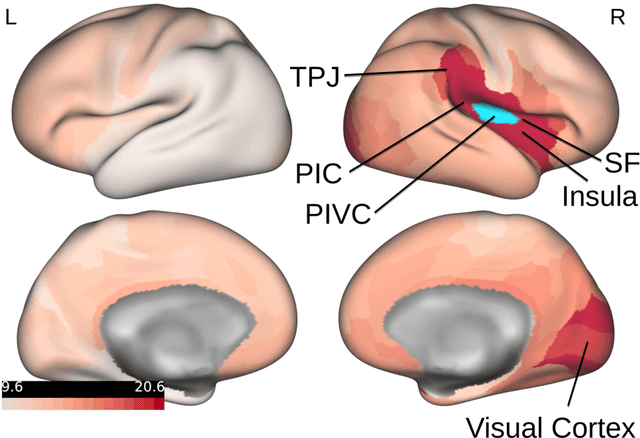
Abstract:A central question in neuroscience is how self-organizing dynamic interactions in the brain emerge on their relatively static structural backbone. Due to the complexity of spatial and temporal dependencies between different brain areas, fully comprehending the interplay between structure and function is still challenging and an area of intense research. In this paper we present a graph neural network (GNN) framework, to describe functional interactions based on the structural anatomical layout. A GNN allows us to process graph-structured spatio-temporal signals, providing a possibility to combine structural information derived from diffusion tensor imaging (DTI) with temporal neural activity profiles, like observed in functional magnetic resonance imaging (fMRI). Moreover, dynamic interactions between different brain regions learned by this data-driven approach can provide a multi-modal measure of causal connectivity strength. We assess the proposed model's accuracy by evaluating its capabilities to replicate empirically observed neural activation profiles, and compare the performance to those of a vector auto regression (VAR), like typically used in Granger causality. We show that GNNs are able to capture long-term dependencies in data and also computationally scale up to the analysis of large-scale networks. Finally we confirm that features learned by a GNN can generalize across MRI scanner types and acquisition protocols, by demonstrating that the performance on small datasets can be improved by pre-training the GNN on data from an earlier and different study. We conclude that the proposed multi-modal GNN framework can provide a novel perspective on the structure-function relationship in the brain. Therewith this approach can be promising for the characterization of the information flow in brain networks.
A constrained ICA-EMD Model for Group Level fMRI Analysis
Mar 22, 2019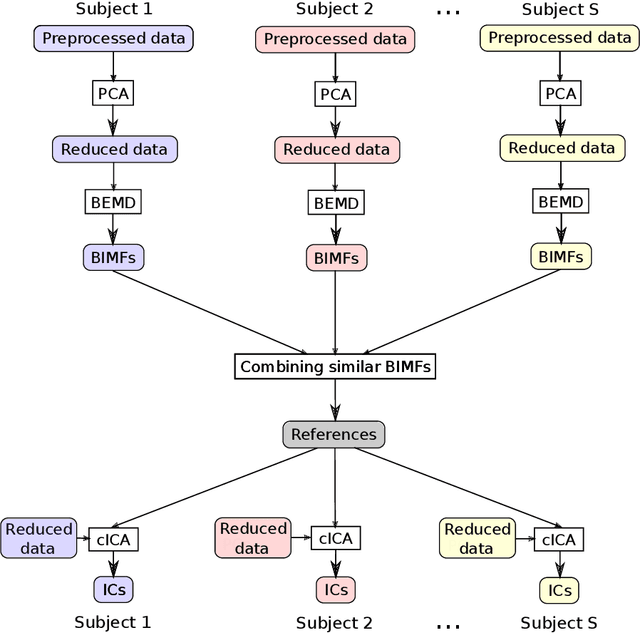
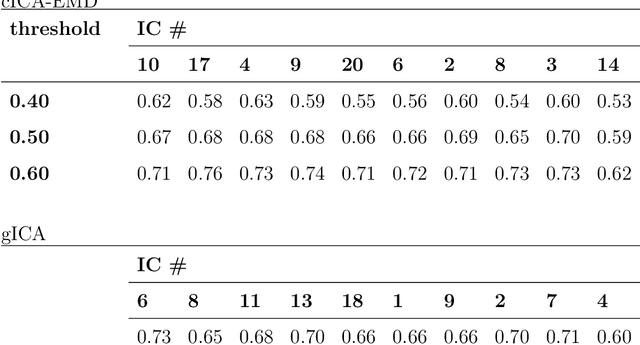


Abstract:Independent component analysis (ICA), as a data driven method, has shown to be a powerful tool for functional magnetic resonance imaging (fMRI) data analysis. One drawback of this multivariate approach is, that it is not compatible to the analysis of group data in general. Therefore various techniques have been proposed in order to overcome this limitation of ICA. In this paper a novel ICA-based work-flow for extracting resting state networks from fMRI group studies is proposed. An empirical mode decomposition (EMD) is used to generate reference signals in a data driven manner, which can be incorporated into a constrained version of ICA (cICA), what helps to eliminate the inherent ambiguities of ICA. The results of the proposed workflow are then compared to those obtained by a widely used group ICA approach for fMRI analysis. In this paper it is demonstrated that intrinsic modes, extracted by EMD, are suitable to serve as references for cICA to obtain typical resting state patterns, which are consistent over subjects. By introducing these reference signals into the ICA, our processing pipeline makes it transparent for the user, how comparable activity patterns across subjects emerge. This additionally allows adapting the trade-off between enforcing similarity across subjects and preserving individual subject features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge