Satya Rajendra Singh
Frequency Disentangled Residual Network
Sep 26, 2021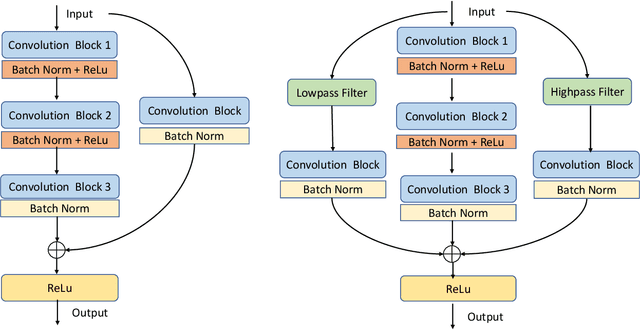
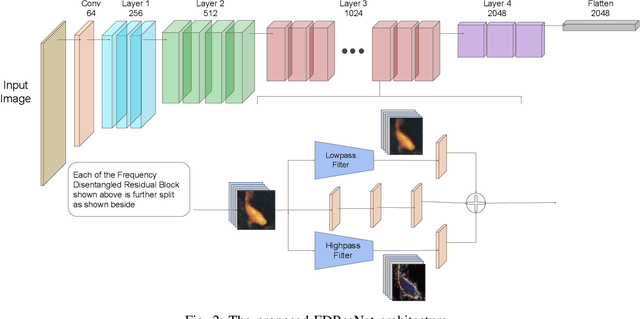
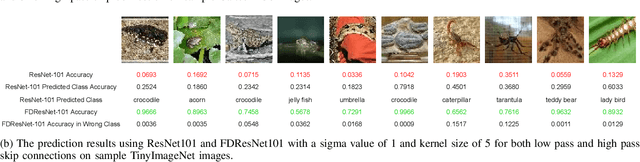
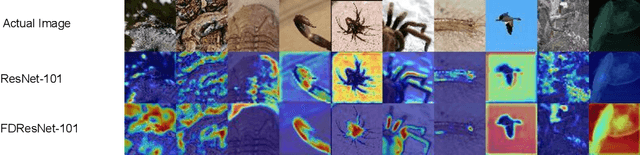
Abstract:Residual networks (ResNets) have been utilized for various computer vision and image processing applications. The residual connection improves the training of the network with better gradient flow. A residual block consists of few convolutional layers having trainable parameters, which leads to overfitting. Moreover, the present residual networks are not able to utilize the high and low frequency information suitably, which also challenges the generalization capability of the network. In this paper, a frequency disentangled residual network (FDResNet) is proposed to tackle these issues. Specifically, FDResNet includes separate connections in the residual block for low and high frequency components, respectively. Basically, the proposed model disentangles the low and high frequency components to increase the generalization ability. Moreover, the computation of low and high frequency components using fixed filters further avoids the overfitting. The proposed model is tested on benchmark CIFAR10/100, Caltech and TinyImageNet datasets for image classification. The performance of the proposed model is also tested in image retrieval framework. It is noticed that the proposed model outperforms its counterpart residual model. The effect of kernel size and standard deviation is also evaluated. The impact of the frequency disentangling is also analyzed using saliency map.
Joint Triplet Autoencoder for Histopathological Colon Cancer Nuclei Retrieval
May 24, 2021



Abstract:Deep learning has shown a great improvement in the performance of visual tasks. Image retrieval is the task of extracting the visually similar images from a database for a query image. The feature matching is performed to rank the images. Various hand-designed features have been derived in past to represent the images. Nowadays, the power of deep learning is being utilized for automatic feature learning from data in the field of biomedical image analysis. Autoencoder and Siamese networks are two deep learning models to learn the latent space (i.e., features or embedding). Autoencoder works based on the reconstruction of the image from latent space. Siamese network utilizes the triplets to learn the intra-class similarity and inter-class dissimilarity. Moreover, Autoencoder is unsupervised, whereas Siamese network is supervised. We propose a Joint Triplet Autoencoder Network (JTANet) by facilitating the triplet learning in autoencoder framework. A joint supervised learning for Siamese network and unsupervised learning for Autoencoder is performed. Moreover, the Encoder network of Autoencoder is shared with Siamese network and referred as the Siamcoder network. The features are extracted by using the trained Siamcoder network for retrieval purpose. The experiments are performed over Histopathological Routine Colon Cancer dataset. We have observed the promising performance using the proposed JTANet model against the Autoencoder and Siamese models for colon cancer nuclei retrieval in histopathological images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge