Satoru Hiwa
Optimized Drug Design using Multi-Objective Evolutionary Algorithms with SELFIES
May 01, 2024Abstract:Computer aided drug design is a promising approach to reduce the tremendous costs, i.e. time and resources, for developing new medicinal drugs. It finds application in aiding the traversal of the vast chemical space of potentially useful compounds. In this paper, we deploy multi-objective evolutionary algorithms, namely NSGA-II, NSGA-III, and MOEA/D, for this purpose. At the same time, we used the SELFIES string representation method. In addition to the QED and SA score, we optimize compounds using the GuacaMol benchmark multi-objective task sets. Our results indicate that all three algorithms show converging behavior and successfully optimize the defined criteria whilst differing mainly in the number of potential solutions found. We observe that novel and promising candidates for synthesis are discovered among obtained compounds in the Pareto-sets.
Generating corneal panoramic images from contact specular microscope images
Jan 06, 2023Abstract:The contact specular microscope has a wider angle of view than that of the non-contact specular microscope but still cannot capture an image of the entire cornea. To obtain such an image, it is necessary to prepare film on the parts of the image captured sequentially and combine them to create a complete image. This study proposes a framework to automatically generate an entire corneal image from videos captured using a contact specular microscope. Relatively focused images were extracted from the videos and panoramic compositing was performed. If an entire image can be generated, it is possible to detect guttae from the image and examine the extent of their presence. The system was implemented and the effectiveness of the proposed framework was examined. The system was implemented using custom-made composite software, Image Composite Software (ICS, K.I. Technology Co., Ltd., Japan, internal algorithms not disclosed), and a supervised learning model using U-Net was used for guttae detection. Several images were correctly synthesized when the constructed system was applied to 94 different corneal videos obtained from Fuchs endothelial corneal dystrophy (FECD) mouse model. The implementation and application of the method to the data in this study confirmed its effectiveness. Owing to the minimal quantitative evaluation performed, such as accuracy with implementation, it may pose some limitations for future investigations.
Deep-learning models in medical image analysis: Detection of esophagitis from the Kvasir Dataset
Jan 06, 2023


Abstract:Early detection of esophagitis is important because this condition can progress to cancer if left untreated. However, the accuracies of different deep learning models in detecting esophagitis have yet to be compared. Thus, this study aimed to compare the accuracies of convolutional neural network models (GoogLeNet, ResNet-50, MobileNet V2, and MobileNet V3) in detecting esophagitis from the open Kvasir dataset of endoscopic images. Results showed that among the models, GoogLeNet achieved the highest F1-scores. Based on the average of true positive rate, MobileNet V3 predicted esophagitis more confidently than the other models. The results obtained using the models were also compared with those obtained using SHapley Additive exPlanations and Gradient-weighted Class Activation Mapping.
Performance Comparison of Deep Learning Architectures for Artifact Removal in Gastrointestinal Endoscopic Imaging
Jan 01, 2022
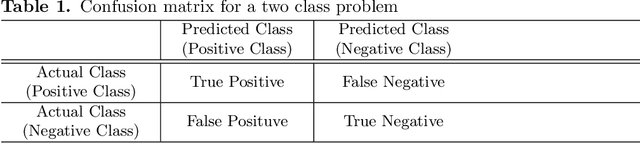
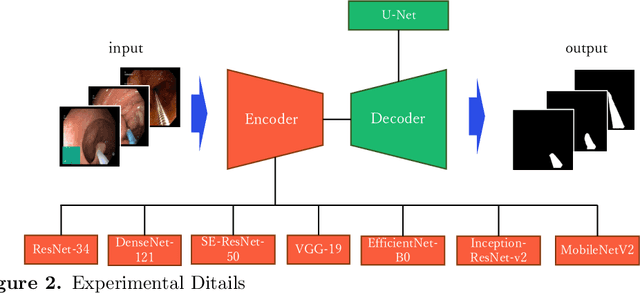
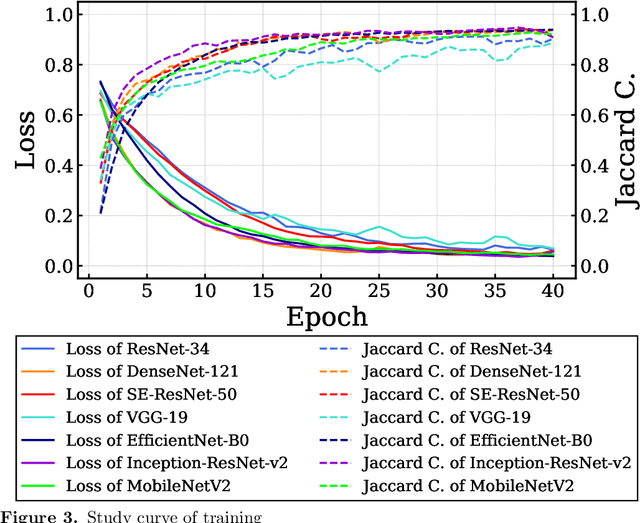
Abstract:Endoscopic images typically contain several artifacts. The artifacts significantly impact image analysis result in computer-aided diagnosis. Convolutional neural networks (CNNs), a type of deep learning, can removes such artifacts. Various architectures have been proposed for the CNNs, and the accuracy of artifact removal varies depending on the choice of architecture. Therefore, it is necessary to determine the artifact removal accuracy, depending on the selected architecture. In this study, we focus on endoscopic surgical instruments as artifacts, and determine and discuss the artifact removal accuracy using seven different CNN architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge