Sébastien Benzekry
ROOFS: RObust biOmarker Feature Selection
Jan 08, 2026Abstract:Feature selection (FS) is essential for biomarker discovery and in the analysis of biomedical datasets. However, challenges such as high-dimensional feature space, low sample size, multicollinearity, and missing values make FS non-trivial. Moreover, FS performances vary across datasets and predictive tasks. We propose roofs, a Python package available at https://gitlab.inria.fr/compo/roofs, designed to help researchers in the choice of FS method adapted to their problem. Roofs benchmarks multiple FS methods on the user's data and generates reports that summarize a comprehensive set of evaluation metrics, including downstream predictive performance estimated using optimism correction, stability, reliability of individual features, and true positive and false positive rates assessed on semi-synthetic data with a simulated outcome. We demonstrate the utility of roofs on data from the PIONeeR clinical trial, aimed at identifying predictors of resistance to anti-PD-(L)1 immunotherapy in lung cancer. The PIONeeR dataset contained 374 multi-source blood and tumor biomarkers from 435 patients. A reduced subset of 214 features was obtained through iterative variance inflation factor pre-filtering. Of the 34 FS methods gathered in roofs, we evaluated 23 in combination with 11 classifiers (253 models in total) and identified a filter based on the union of Benjamini-Hochberg false discovery rate-adjusted p-values from t-test and logistic regression as the optimal approach, outperforming other methods including the widely used LASSO. We conclude that comprehensive benchmarking with roofs has the potential to improve the robustness and reproducibility of FS discoveries and increase the translational value of clinical models.
Une comparaison des algorithmes d'apprentissage pour la survie avec données manquantes
Mar 23, 2023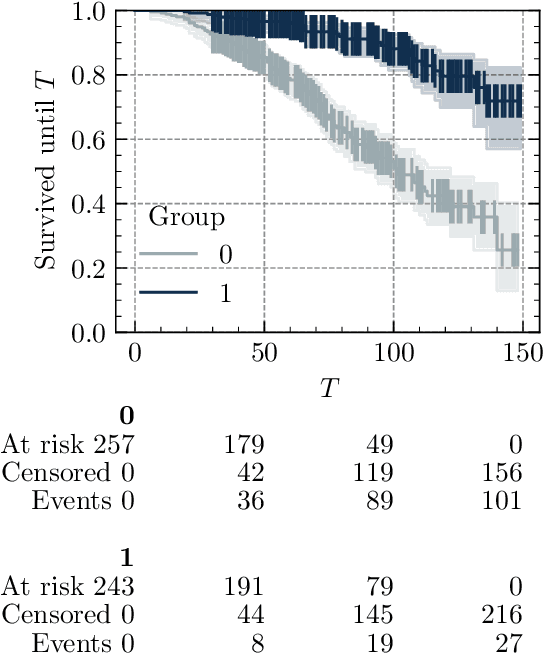
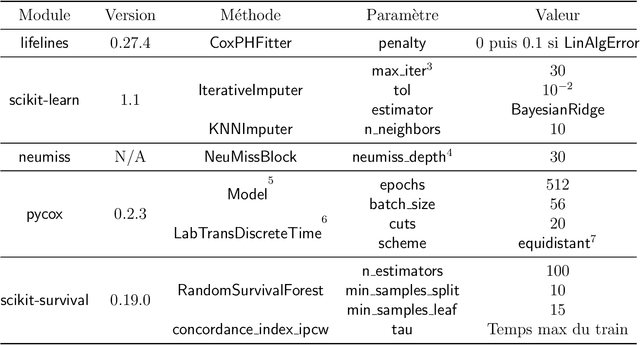
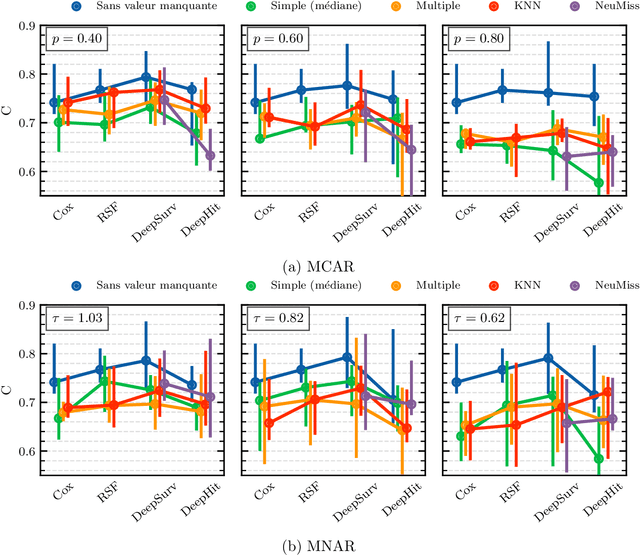
Abstract:Survival analysis is an essential tool for the study of health data. An inherent component of such data is the presence of missing values. In recent years, researchers proposed new learning algorithms for survival tasks based on neural networks. Here, we studied the predictive performance of such algorithms coupled with different methods for handling missing values on simulated data that reflect a realistic situation, i.e., when individuals belong to unobserved clusters. We investigated different patterns of missing data. The results show that, without further feature engineering, no single imputation method is better than the others in all cases. The proposed methodology can be used to compare other missing data patterns and/or survival models. The Python code is accessible via the package survivalsim. -- L'analyse de survie est un outil essentiel pour l'\'etude des donn\'ees de sant\'e. Une composante inh\'erente \`a ces donn\'ees est la pr\'esence de valeurs manquantes. Ces derni\`eres ann\'ees, de nouveaux algorithmes d'apprentissage pour la survie, bas\'es sur les r\'eseaux de neurones, ont \'et\'e con\c{c}us. L'objectif de ce travail est d'\'etudier la performance en pr\'ediction de ces algorithmes coupl\'es \`a diff\'erentes m\'ethodes pour g\'erer les valeurs manquantes, sur des donn\'ees simul\'ees qui refl\`etent une situation rencontr\'ee en pratique, c'est-\`a dire lorsque les individus peuvent \^etre group\'es selon leurs covariables. Diff\'erents sch\'emas de donn\'ees manquantes sont \'etudi\'es. Les r\'esultats montrent que, sans l'ajout de variables suppl\'ementaires, aucune m\'ethode d'imputation n'est meilleure que les autres dans tous les cas. La m\'ethodologie propos\'ee peut \^etre utilis\'ee pour comparer d'autres mod\`eles de survie. Le code en Python est accessible via le package survivalsim.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge