Robert O'Shea
Closed-Form Feedback-Free Learning with Forward Projection
Jan 27, 2025Abstract:State-of-the-art methods for backpropagation-free learning employ local error feedback to direct iterative optimisation via gradient descent. In this study, we examine the more restrictive setting where retrograde communication from neuronal outputs is unavailable for pre-synaptic weight optimisation. To address this challenge, we propose Forward Projection (FP). This novel randomised closed-form training method requires only a single forward pass over the entire dataset for model fitting, without retrograde communication. Target values for pre-activation membrane potentials are generated layer-wise via nonlinear projections of pre-synaptic inputs and the labels. Local loss functions are optimised over pre-synaptic inputs using closed-form regression, without feedback from neuronal outputs or downstream layers. Interpretability is a key advantage of FP training; membrane potentials of hidden neurons in FP-trained networks encode information which is interpretable layer-wise as label predictions. We demonstrate the effectiveness of FP across four biomedical datasets. In few-shot learning tasks, FP yielded more generalisable models than those optimised via backpropagation. In large-sample tasks, FP-based models achieve generalisation comparable to gradient descent-based local learning methods while requiring only a single forward propagation step, achieving significant speed up for training. Interpretation functions defined on local neuronal activity in FP-based models successfully identified clinically salient features for diagnosis in two biomedical datasets. Forward Projection is a computationally efficient machine learning approach that yields interpretable neural network models without retrograde communication of neuronal activity during training.
Gextext: Disease Network Extraction from Biomedical Literature
Dec 17, 2019
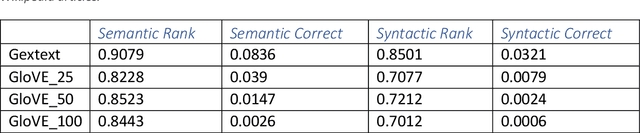


Abstract:PURPOSE: We propose a fully unsupervised method to learn latent disease networks directly from unstructured biomedical text corpora. This method addresses current challenges in unsupervised knowledge extraction, such as the detection of long-range dependencies and requirements for large training corpora. METHODS: Let C be a corpus of n text chunks. Let V be a set of p disease terms occurring in the corpus. Let X indicate the occurrence of V in C. Gextext identifies disease similarities by positively correlated occurrence patterns. This information is combined to generate a graph on which geodesic distance describes dissimilarity. Diseasomes were learned by Gextext and GloVE on corpora of 100-1000 PubMed abstracts. Similarity matrix estimates were validated against biomedical semantic similarity metrics and gene profile similarity. RESULTS: Geodesic distance on Gextext-inferred diseasomes correlated inversely with external measures of semantic similarity. Gene profile similarity also correlated significant with proximity on the inferred graph. Gextext outperformed GloVE in our experiments. The information contained on the Gextext graph exceeded the explicit information content within the text. CONCLUSIONS: Gextext extracts latent relationships from unstructured text, enabling fully unsupervised modelling of diseasome graphs from PubMed abstracts.
Model Selection With Graphical Neighbour Information
Aug 27, 2019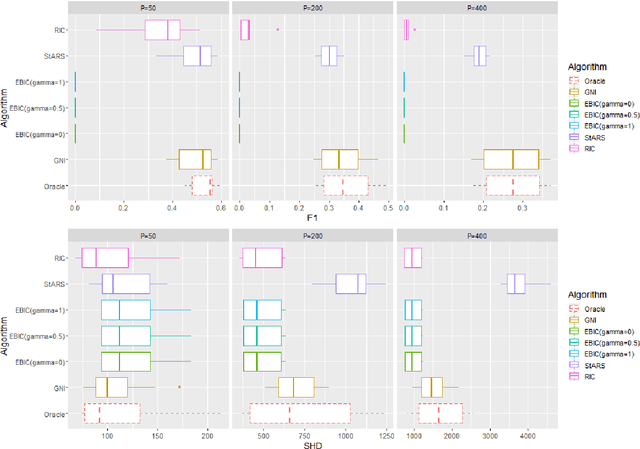
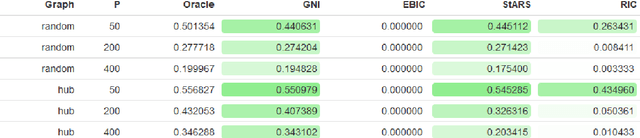
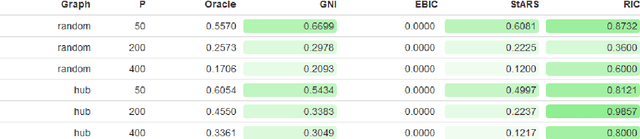
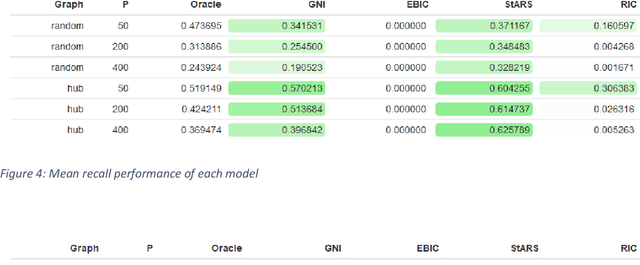
Abstract:Accurate model selection is a fundamental requirement for statistical analysis. In many real-world applications of graphical modelling, correct model structure identification is the ultimate objective. Standard model validation procedures such as information theoretic scores and cross validation have demonstrated poor performance in the high dimensional setting. Specialised methods such as EBIC, StARS and RIC have been developed for the explicit purpose of high-dimensional Gaussian graphical model selection. We present a novel model score criterion, Graphical Neighbour Information. This method demonstrates oracle performance in high-dimensional model selection, outperforming the current state-of-the-art in our simulations. The Graphical Neighbour Information criterion has the additional advantage of efficient, closed-form computability, sparing the costly inference of multiple models on data subsamples. We provide a theoretical analysis of the method and benchmark simulations versus the current state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge