Pedro Seber
Predicting O-GlcNAcylation Sites in Mammalian Proteins with Transformers and RNNs Trained with a New Loss Function
Feb 27, 2024Abstract:Glycosylation, a protein modification, has multiple essential functional and structural roles. O-GlcNAcylation, a subtype of glycosylation, has the potential to be an important target for therapeutics, but methods to reliably predict O-GlcNAcylation sites had not been available until 2023; a 2021 review correctly noted that published models were insufficient and failed to generalize. Moreover, many are no longer usable. In 2023, a considerably better RNN model with an F$_1$ score of 36.17% and an MCC of 34.57% on a large dataset was published. This article first sought to improve these metrics using transformer encoders. While transformers displayed high performance on this dataset, their performance was inferior to that of the previously published RNN. We then created a new loss function, which we call the weighted focal differentiable MCC, to improve the performance of classification models. RNN models trained with this new function display superior performance to models trained using the weighted cross-entropy loss; this new function can also be used to fine-tune trained models. A two-cell RNN trained with this loss achieves state-of-the-art performance in O-GlcNAcylation site prediction with an F$_1$ score of 38.82% and an MCC of 38.21% on that large dataset.
LCEN: A Novel Feature Selection Algorithm for Nonlinear, Interpretable Machine Learning Models
Feb 27, 2024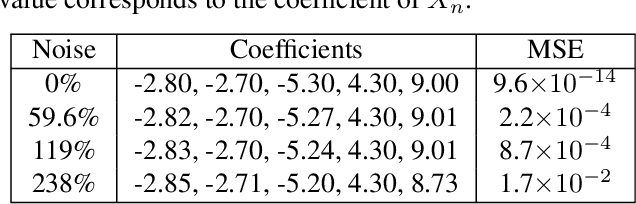
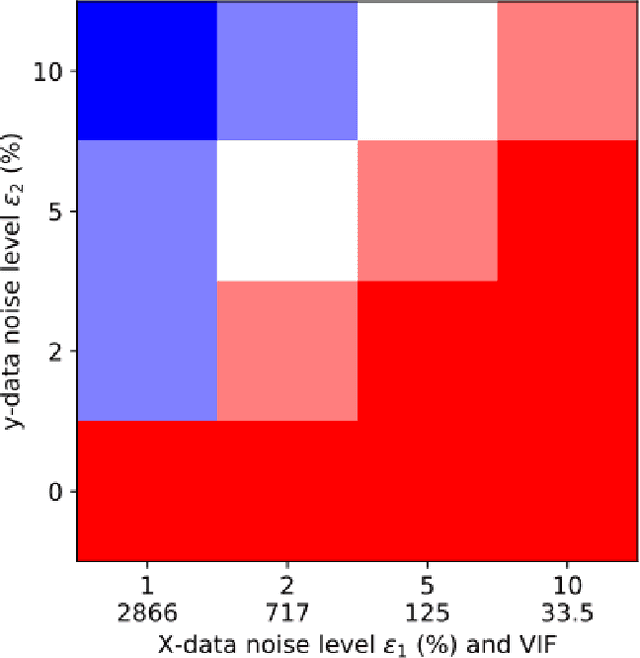
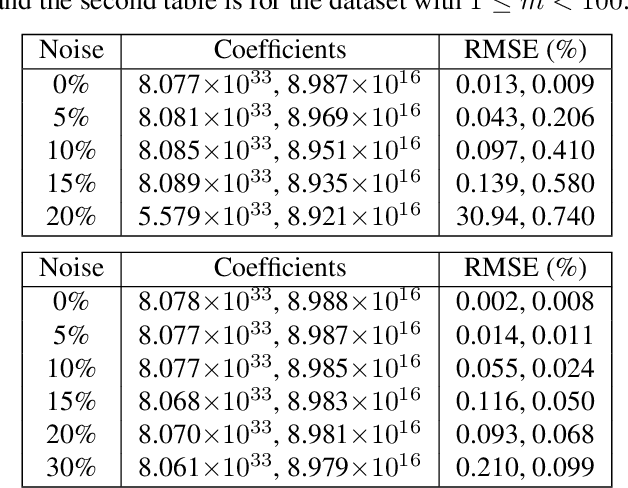
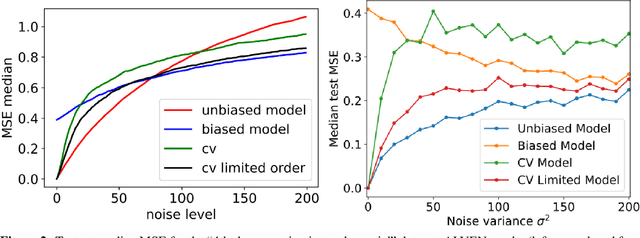
Abstract:Interpretable architectures can have advantages over black-box architectures, and interpretability is essential for the application of machine learning in critical settings, such as aviation or medicine. However, the simplest, most commonly used interpretable architectures (such as LASSO or EN) are limited to linear predictions and have poor feature selection capabilities. In this work, we introduce the LASSO-Clip-EN (LCEN) algorithm for the creation of nonlinear, interpretable machine learning models. LCEN is tested on a wide variety of artificial and empirical datasets, creating more accurate, sparser models than other commonly used architectures. These experiments reveal that LCEN is robust against many issues typically present in datasets and modeling, including noise, multicollinearity, data scarcity, and hyperparameter variance. LCEN is also able to rediscover multiple physical laws from empirical data and, for processes with no known physical laws, LCEN achieves better results than many other dense and sparse methods -- including using 10.8 times fewer features than dense methods and 8.1 times fewer features than EN on one dataset, and is comparable to an ANN on another dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge