N. Lin
A General Statistic Framework for Genome-based Disease Risk Prediction
Oct 27, 2014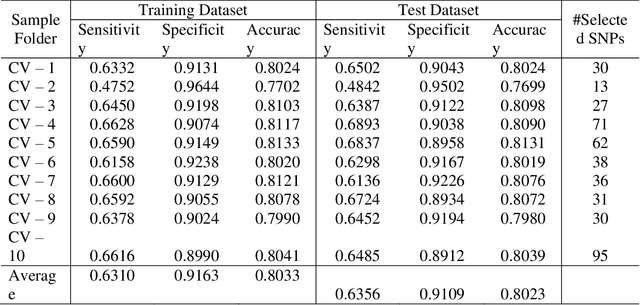
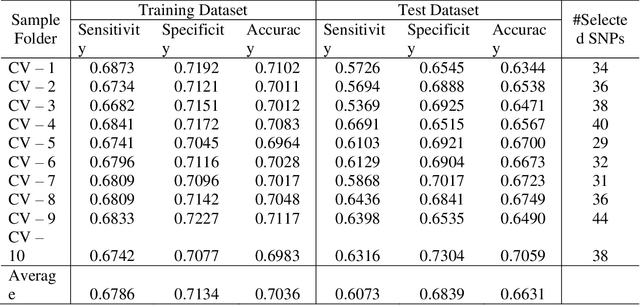
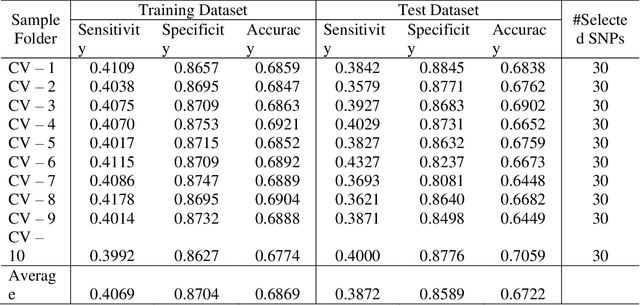
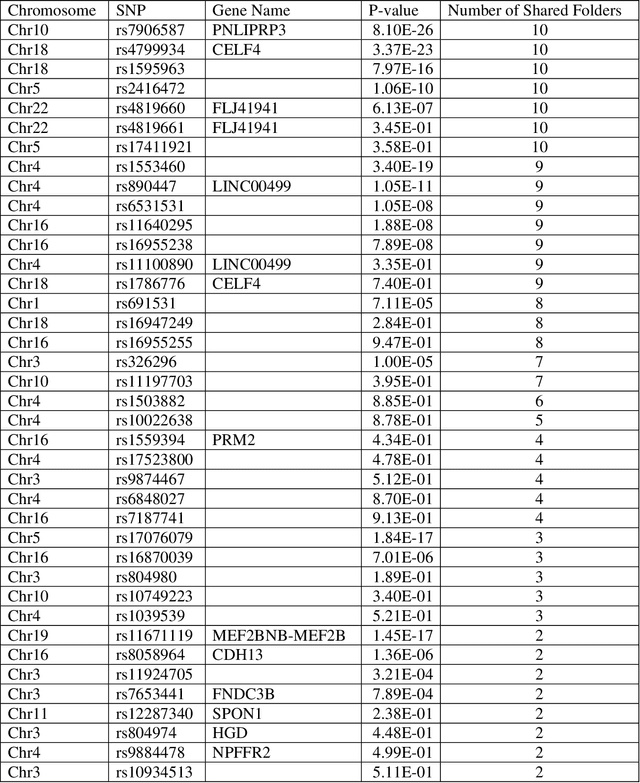
Abstract:Advances of modern sensing and sequencing technologies generate a deluge of high dimensional space-temporal physiological and next-generation sequencing (NGS) data. Physiological traits are observed either as continuous random functions, or on a dense grid and referred to as function-valued traits. Both physiological and NGS data are highly correlated data with their inherent order, spacing, and functional nature which are ignored by traditional summary-based univariate and multivariate regression methods designed for quantitative genetic analysis of scalar trait and common variants. To capture morphological and dynamic features of the data and utilize their dependent structure, we propose a functional linear model (FLM) in which a trait curve is modeled as a response function, the genetic variation in a genomic region or gene is modeled as a functional predictor, and the genetic effects are modeled as a function of both time and genomic position (FLMF) for genetic analysis of function-valued trait with both GWAS and NGS data. By extensive simulations, we demonstrate that the FLMF has the correct type 1 error rates and much higher power to detect association than the existing methods. The FLMF is applied to sleep data from Starr County health studies where oxygen saturation were measured in 22,670 seconds on average for 833 individuals. We found 65 genes that were significantly associated with oxygen saturation functional trait with P-values ranging from 2.40E-06 to 2.53E-21. The results clearly demonstrate that the FLMF substantially outperforms the traditional genetic models with scalar trait.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge