Get our free extension to see links to code for papers anywhere online!Free add-on: code for papers everywhere!Free add-on: See code for papers anywhere!
Miklos Csuros
Provably Fast and Accurate Recovery of Evolutionary Trees through Harmonic Greedy Triplets
Nov 23, 2000Figures and Tables: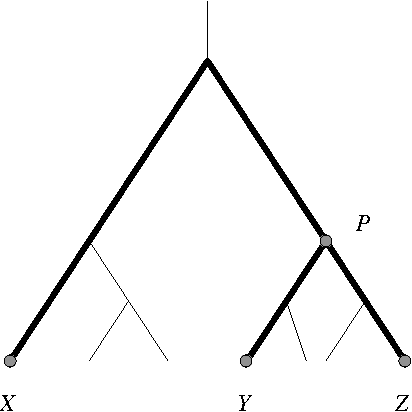
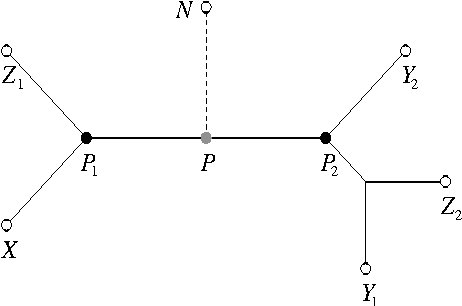
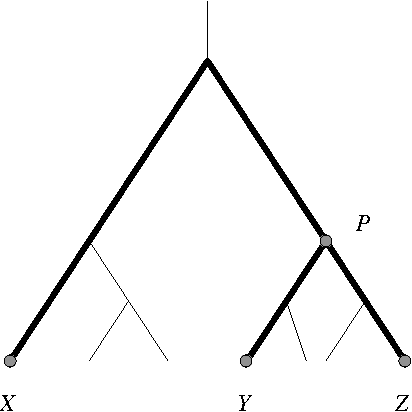

Abstract:We give a greedy learning algorithm for reconstructing an evolutionary tree based on a certain harmonic average on triplets of terminal taxa. After the pairwise distances between terminal taxa are estimated from sequence data, the algorithm runs in O(n^2) time using O(n) work space, where n is the number of terminal taxa. These time and space complexities are optimal in the sense that the size of an input distance matrix is n^2 and the size of an output tree is n. Moreover, in the Jukes-Cantor model of evolution, the algorithm recovers the correct tree topology with high probability using sample sequences of length polynomial in (1) n, (2) the logarithm of the error probability, and (3) the inverses of two small parameters.
* The paper will appear in SIAM Journal on Computing
Via
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge