Matthew Guy
Flexible Blood Glucose Control: Offline Reinforcement Learning from Human Feedback
Jan 27, 2025



Abstract:Reinforcement learning (RL) has demonstrated success in automating insulin dosing in simulated type 1 diabetes (T1D) patients but is currently unable to incorporate patient expertise and preference. This work introduces PAINT (Preference Adaptation for INsulin control in T1D), an original RL framework for learning flexible insulin dosing policies from patient records. PAINT employs a sketch-based approach for reward learning, where past data is annotated with a continuous reward signal to reflect patient's desired outcomes. Labelled data trains a reward model, informing the actions of a novel safety-constrained offline RL algorithm, designed to restrict actions to a safe strategy and enable preference tuning via a sliding scale. In-silico evaluation shows PAINT achieves common glucose goals through simple labelling of desired states, reducing glycaemic risk by 15% over a commercial benchmark. Action labelling can also be used to incorporate patient expertise, demonstrating an ability to pre-empt meals (+10% time-in-range post-meal) and address certain device errors (-1.6% variance post-error) with patient guidance. These results hold under realistic conditions, including limited samples, labelling errors, and intra-patient variability. This work illustrates PAINT's potential in real-world T1D management and more broadly any tasks requiring rapid and precise preference learning under safety constraints.
The Safety Challenges of Deep Learning in Real-World Type 1 Diabetes Management
Oct 23, 2023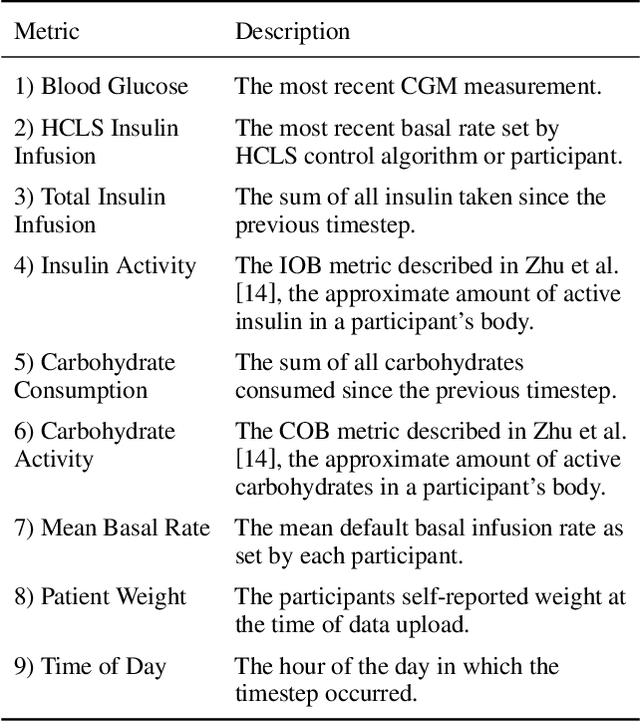
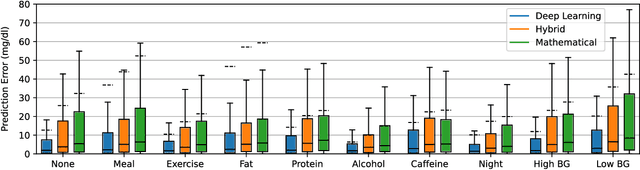
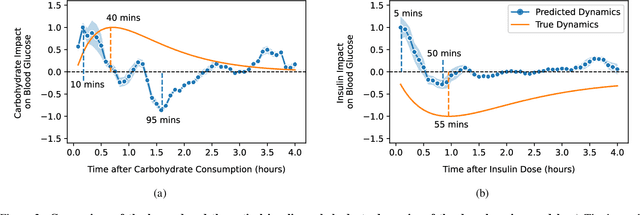
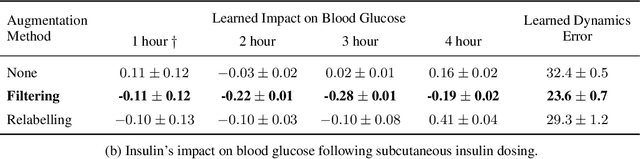
Abstract:Blood glucose simulation allows the effectiveness of type 1 diabetes (T1D) management strategies to be evaluated without patient harm. Deep learning algorithms provide a promising avenue for extending simulator capabilities; however, these algorithms are limited in that they do not necessarily learn physiologically correct glucose dynamics and can learn incorrect and potentially dangerous relationships from confounders in training data. This is likely to be more important in real-world scenarios, as data is not collected under strict research protocol. This work explores the implications of using deep learning algorithms trained on real-world data to model glucose dynamics. Free-living data was processed from the OpenAPS Data Commons and supplemented with patient-reported tags of challenging diabetes events, constituting one of the most detailed real-world T1D datasets. This dataset was used to train and evaluate state-of-the-art glucose simulators, comparing their prediction error across safety critical scenarios and assessing the physiological appropriateness of the learned dynamics using Shapley Additive Explanations (SHAP). While deep learning prediction accuracy surpassed the widely-used mathematical simulator approach, the model deteriorated in safety critical scenarios and struggled to leverage self-reported meal and exercise information. SHAP value analysis also indicated the model had fundamentally confused the roles of insulin and carbohydrates, which is one of the most basic T1D management principles. This work highlights the importance of considering physiological appropriateness when using deep learning to model real-world systems in T1D and healthcare more broadly, and provides recommendations for building models that are robust to real-world data constraints.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge